1BQS
 
 | | THE CRYSTAL STRUCTURE OF MUCOSAL ADDRESSIN CELL ADHESION MOLECULE-1 (MADCAM-1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (MUCOSAL ADDRESSIN CELL ADHESION MOLECULE-1) | | Authors: | Tan, K, Casasnovas, J.M, Liu, J.H, Briskin, M.J, Springer, T.A, Wang, J.-H. | | Deposit date: | 1998-08-18 | | Release date: | 1999-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of immunoglobulin superfamily domains 1 and 2 of MAdCAM-1 reveals novel features important for integrin recognition.
Structure, 6, 1998
|
|
1MV8
 
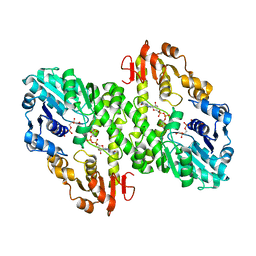 | | 1.55 A crystal structure of a ternary complex of GDP-mannose dehydrogenase from Psuedomonas aeruginosa | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, GDP-mannose 6-dehydrogenase, ... | | Authors: | Snook, C.F, Tipton, P.A, Beamer, L.J. | | Deposit date: | 2002-09-24 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of GDP-mannose
dehydrogenase: A key enzyme in alginate
biosynthesis of P. aeruginosa
Biochemistry, 42, 2003
|
|
1C4E
 
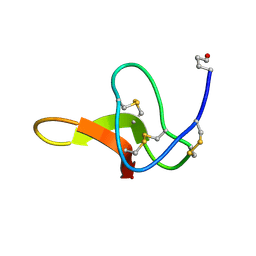 | |
3HP6
 
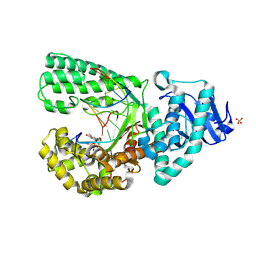 | |
4XRX
 
 | | Crystal structure of a metabolic reductase with (E)-5-((1-methyl-5-oxo-2-thioxoimidazolidin-4-ylidene)methyl)pyridin-2(1H)-one | | Descriptor: | 5-[(E)-(1-methyl-5-oxo-2-thioxoimidazolidin-4-ylidene)methyl]pyridin-2(1H)-one, Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, B, Wu, F, Jiang, H, Kogiso, M, Yao, Y, Zhou, C, Li, X, Song, Y. | | Deposit date: | 2015-01-21 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Inhibition of Cancer-Associated Mutant Isocitrate Dehydrogenases by 2-Thiohydantoin Compounds.
J.Med.Chem., 58, 2015
|
|
1C4O
 
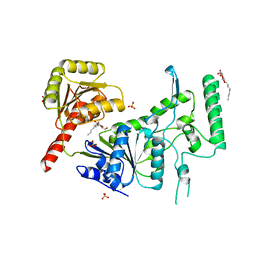 | | CRYSTAL STRUCTURE OF THE DNA NUCLEOTIDE EXCISION REPAIR ENZYME UVRB FROM THERMUS THERMOPHILUS | | Descriptor: | DNA NUCLEOTIDE EXCISION REPAIR ENZYME UVRB, SULFATE ION, octyl beta-D-glucopyranoside | | Authors: | Machius, M, Henry, L, Palnitkar, M, Deisenhofer, J. | | Deposit date: | 1999-09-14 | | Release date: | 2000-07-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the DNA nucleotide excision repair enzyme UvrB from Thermus thermophilus.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3HZV
 
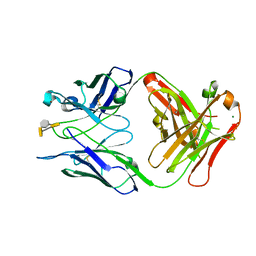 | | Crystal structure of S73-2 antibody in complex with antigen Kdo(2.8)Kdo(2.4)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, MAGNESIUM ION, S73-2 Fab (IgG1k) heavy chain, ... | | Authors: | Brooks, C.L, Muller-Loennies, S, Borisova, S.N, Brade, L, Kosma, P, Hirama, T, MacKenzie, C.R, Brade, H, Evans, S.V. | | Deposit date: | 2009-06-24 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibodies raised against chlamydial lipopolysaccharide antigens reveal convergence in germline gene usage and differential epitope recognition
Biochemistry, 49, 2010
|
|
8E1W
 
 | | Neutron crystal structure of Panus similis AA9A at room temperature | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Meilleur, F, Tandrup, T, Lo Leggio, L. | | Deposit date: | 2022-08-11 | | Release date: | 2023-01-11 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2.1 Å), X-RAY DIFFRACTION | | Cite: | Joint X-ray/neutron structure of Lentinus similis AA9_A at room temperature.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
3JWO
 
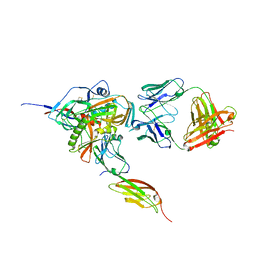 | | Structure of HIV-1 gp120 with gp41-Interactive Region: Layered Architecture and Basis of Conformational Mobility | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB 48D Heavy CHAIN, FAB 48D LIGHT CHAIN, ... | | Authors: | Pancera, M, Majeed, S, Huang, C.C, Kwon, Y.D, Zhou, T, Robinson, J.E, Sodroski, J, Wyatt, R, Kwong, P.D. | | Deposit date: | 2009-09-18 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6W35
 
 | |
8G1A
 
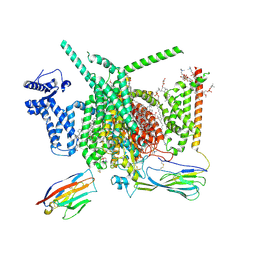 | | Cryo-EM structure of Nav1.7 with CBD | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-02-01 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cannabidiol inhibits Na v channels through two distinct binding sites.
Nat Commun, 14, 2023
|
|
3JZJ
 
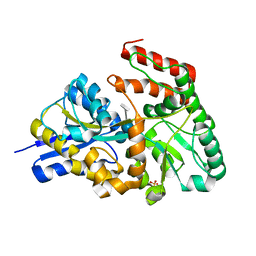 | | Crystal structures of the GacH receptor of Streptomyces glaucescens GLA.O in the unliganded form and in complex with acarbose and an acarbose homolog. Comparison with acarbose-loaded maltose binding protein of Salmonella typhimurium. | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Acarbose/maltose binding protein GacH, SULFATE ION | | Authors: | Vahedi-Faridi, A, Licht, A, Bulut, H, Schneider, E. | | Deposit date: | 2009-09-23 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of the Solute Receptor GacH of Streptomyces glaucescens in Complex with Acarbose and an Acarbose Homolog: Comparison with the Acarbose-Loaded Maltose-Binding Protein of Salmonella typhimurium.
J.Mol.Biol., 397, 2010
|
|
6NMY
 
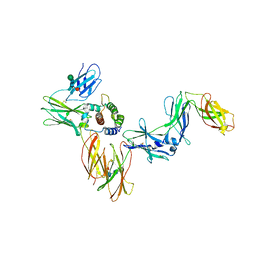 | | A Cytokine-receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit beta, ... | | Authors: | Dhagat, U, Kan, W.L, Hercus, T.R, Broughton, S.E, Nero, T.L, Lopez, A.F, Parker, M.W. | | Deposit date: | 2019-01-13 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Signalling conformation of cell surface receptor
To Be Published
|
|
8U28
 
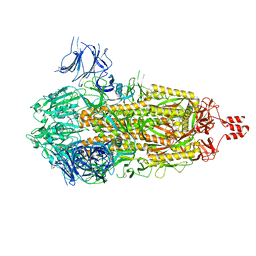 | |
8TYP
 
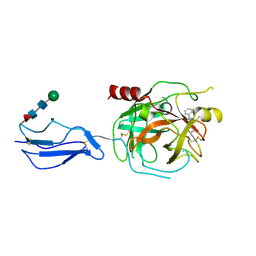 | | Complement Protease C1s Inhibited by 6-(4-phenylpiperazin-1-yl)pyridine-3-carboximidamide | | Descriptor: | 6-(4-phenylpiperazin-1-yl)pyridine-3-carboximidamide, Complement C1s subcomponent, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Geisbrecht, B.V. | | Deposit date: | 2023-08-25 | | Release date: | 2023-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of the C1s Protease and the Classical Complement Pathway by 6-(4-Phenylpiperazin-1-yl)Pyridine-3-Carboximidamide and Chemical Analogs.
J Immunol., 212, 2024
|
|
6NVF
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 8 | | Descriptor: | (4-{[(1R,2R)-2-(carboxymethyl)cyclopentyl]methyl}phenyl)acetic acid, (4-{[(1S,2S)-2-(carboxymethyl)cyclopentyl]methyl}phenyl)acetic acid, ATP-dependent dethiobiotin synthetase BioD, ... | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-02-05 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 8
To Be Published
|
|
1QM8
 
 | | Structure of Bacteriorhodopsin at 100 K | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, 3-PHOSPHORYL-[1,2-DI-PHYTANYL]GLYCEROL, ... | | Authors: | Takeda, K, Matsui, Y, Sato, H, Hino, T, Kanamori, E, Okumura, H, Yamane, T, Kamiya, N, Kouyama, T. | | Deposit date: | 1999-09-22 | | Release date: | 2000-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Three-Dimensional Crystal of Bacteriorhodopsin Obtained by Successive Fusion of the Vesicular Assemblies.
J.Mol.Biol., 283, 1998
|
|
8UUL
 
 | | Prototypic SARS-CoV-2 spike (containing K417) in the closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Geng, Q, Liu, B, Li, F. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein.
Elife, 12, 2023
|
|
8UUM
 
 | | Prototypic SARS-CoV-2 spike (containing K417) in the open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose, ... | | Authors: | Geng, Q, Liu, B, Li, F. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein.
Elife, 12, 2023
|
|
8UUN
 
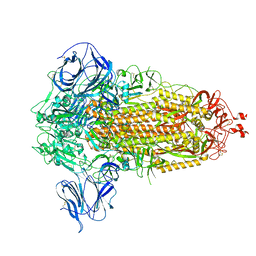 | | Prototypic SARS-CoV-2 spike (containing V417) in the closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Geng, Q, Liu, B, Li, F. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein.
Elife, 12, 2023
|
|
8UUO
 
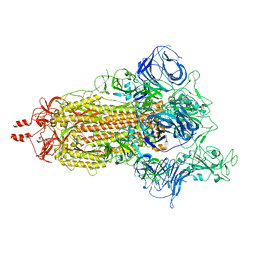 | | Prototypic SARS-CoV-2 spike (containing V417) in the open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose, ... | | Authors: | Geng, Q, Liu, B, Li, F. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein.
Elife, 12, 2023
|
|
6T7H
 
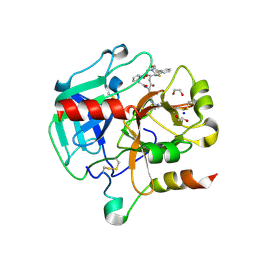 | | Crystal structure of Thrombin in complex with macrocycle N14-PR4-A | | Descriptor: | (14S,17R)-14-(3-carbamimidamidopropyl)-3-(furan-2-ylmethyl)-5,12,15-tris(oxidanylidene)-19-thia-3,6,13,16-tetrazatricyclo[19.4.0.0^{6,10}]pentacosa-1(25),7,9,21,23-pentaene-17-carboxamide, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Angelini, A, Kumar, M.G, Heinis, C, Cendron, L. | | Deposit date: | 2019-10-22 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Macrocycle synthesis strategy based on step-wise "adding and reacting" three components enables screening of large combinatorial libraries.
Chem Sci, 11, 2020
|
|
3HTT
 
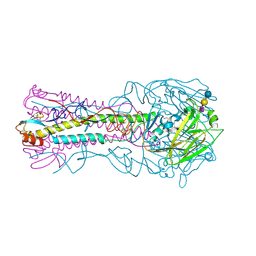 | | The hemagglutinin structure of an avian H1N1 influenza A virus in complex with 2,3-sialyllactose | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Wang, G, Li, A, Zhang, Q, Wu, C, Zhang, R, Cai, Q, Song, W, Yuen, K.-Y. | | Deposit date: | 2009-06-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The hemagglutinin structure of an avian H1N1 influenza A virus
Virology, 392, 2009
|
|
7P32
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with cyclosulfamidate 6 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Kok, K, Overkleeft, H, Artola, M, Sulzenbacher, G. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | 1,6- epi-Cyclophellitol Cyclosulfamidate Is a Bona Fide Lysosomal alpha-Glucosidase Stabilizer for the Treatment of Pompe Disease.
J.Am.Chem.Soc., 144, 2022
|
|
6T7L
 
 | | Crystal structure of AmpC from E.coli with Nacubactam (OP0595) | | Descriptor: | (2S,5R)-N-(2-aminoethoxy)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase, ... | | Authors: | Lang, P.A, Leissing, T.M, Schofield, C.J, Brem, J. | | Deposit date: | 2019-10-22 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Investigations of the Inhibition of Escherichia coli AmpC beta-Lactamase by Diazabicyclooctanes.
Antimicrob.Agents Chemother., 65, 2021
|
|
