231L
 
 | | T4 LYSOZYME MUTANT M106K | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Lipscomb, L.A, Drew, D.L, Gassner, N, Baase, W.A, Matthews, B.W. | | Deposit date: | 1997-10-03 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Context-dependent protein stabilization by methionine-to-leucine substitution shown in T4 lysozyme.
Protein Sci., 7, 1998
|
|
1OYU
 
 | |
171L
 
 | |
1QS5
 
 | |
189L
 
 | |
1QTC
 
 | |
262L
 
 | |
3ODU
 
 | | The 2.5 A structure of the CXCR4 chemokine receptor in complex with small molecule antagonist IT1t | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-27 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
7S3I
 
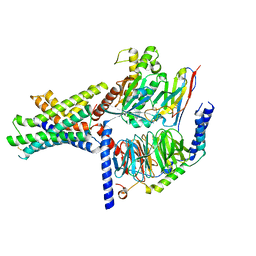 | | Ex4-D-Ala bound to the glucagon-like peptide-1 receptor/g protein complex (conformer 2) | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Belousoff, M.J, Piper, S.J, Danev, R. | | Deposit date: | 2021-09-07 | | Release date: | 2022-01-05 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structural and functional diversity among agonist-bound states of the GLP-1 receptor.
Nat.Chem.Biol., 18, 2022
|
|
6ZX9
 
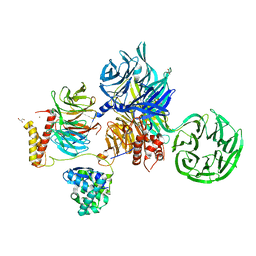 | | Crystal structure of SIV Vpr,fused to T4 lysozyme, isolated from moustached monkey, bound to human DDB1 and human DCAF1 (amino acid residues 1046-1396) | | Descriptor: | DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, GLYCEROL, ... | | Authors: | Schwefel, D, Banchenko, S. | | Deposit date: | 2020-07-29 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.519729 Å) | | Cite: | Structural insights into Cullin4-RING ubiquitin ligase remodelling by Vpr from simian immunodeficiency viruses.
Plos Pathog., 17, 2021
|
|
8WG7
 
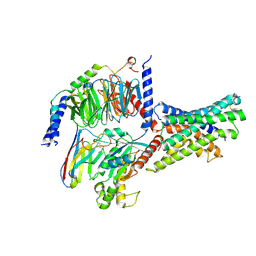 | | Cryo-EM structures of peptide free and Gs-coupled GLP-1R | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cong, Z.T, Zhao, F.H, Li, Y, Luo, G, Zhou, Q.T, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-09-20 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Molecular features of the ligand-free GLP-1R, GCGR and GIPR in complex with G s proteins.
Cell Discov, 10, 2024
|
|
7P2L
 
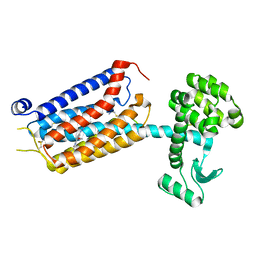 | | thermostabilised 7TM domain of human mGlu5 receptor bound to photoswitchable ligand alloswitch-1 | | Descriptor: | 2-chloranyl-~{N}-[2-methoxy-4-[(~{E})-pyridin-2-yldiazenyl]phenyl]benzamide, Metabotropic glutamate receptor 5,Endolysin,Metabotropic glutamate receptor 5 | | Authors: | Huang, C.Y, Vinothkumar, K.R, Lebon, G. | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Agonists and allosteric modulators promote signaling from different metabotropic glutamate receptor 5 conformations.
Cell Rep, 36, 2021
|
|
2QB0
 
 | |
8JIR
 
 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist SAR425899-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4ARJ
 
 | | Crystal structure of a pesticin (translocation and receptor binding domain) from Y. pestis and T4-lysozyme chimera | | Descriptor: | PESTICIN, LYSOZYME, SULFATE ION | | Authors: | Zeth, K, Patzer, S.I, Albrecht, R, Braun, V. | | Deposit date: | 2012-04-24 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Structure and Mechanistic Studies of Pesticin, a Bacterial Homolog of Phage Lysozymes.
J.Biol.Chem., 287, 2012
|
|
3EML
 
 | | The 2.6 A Crystal Structure of a Human A2A Adenosine Receptor bound to ZM241385. | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Human Adenosine A2A receptor/T4 lysozyme chimera, STEARIC ACID, ... | | Authors: | Jaakola, V.-P, Griffith, M.T, Hanson, M.A, Cherezov, V, Chien, E.Y.T, Lane, J.R, Ijzerman, A.P, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2008-09-24 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The 2.6 Angstrom Crystal Structure of a Human A2A Adenosine Receptor Bound to an Antagonist.
Science, 322, 2008
|
|
8DCR
 
 | | Cryo-EM structure of dobutamine-bound beta1-adrenergic receptor in complex with heterotrimeric Gs-protein | | Descriptor: | DOBUTAMINE, Endolysin,Endolysin,Beta-1 adrenergic receptor chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Su, M, Paknejad, N, Hite, R.K, Huang, X.Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of beta 1 -adrenergic receptor in complex with Gs and ligands of different efficacies.
Nat Commun, 13, 2022
|
|
5EWX
 
 | | Fusion protein of T4 lysozyme and B4 domain of protein A from staphylococcal aureus with chemical cross-linker EY-CBS | | Descriptor: | 2,2'-ethyne-1,2-diylbis{5-[(chloroacetyl)amino]benzenesulfonic acid}, Endolysin,Immunoglobulin G-binding protein A,Endolysin | | Authors: | Jeong, W.H, Lee, H, Song, D.H, Lee, J.O. | | Deposit date: | 2015-11-22 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Connecting two proteins using a fusion alpha helix stabilized by a chemical cross linker.
Nat Commun, 7, 2016
|
|
4OO9
 
 | | Structure of the human class C GPCR metabotropic glutamate receptor 5 transmembrane domain in complex with the negative allosteric modulator mavoglurant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mavoglurant, Metabotropic glutamate receptor 5, ... | | Authors: | Dore, A.S, Okrasa, K, Patel, J.C, Serrano-Vega, M, Bennett, K, Cooke, R.M, Errey, J.C, Jazayeri, A, Khan, S, Tehan, B, Weir, M, Wiggin, G.R, Marshall, F.H. | | Deposit date: | 2014-01-31 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain.
Nature, 511, 2014
|
|
5WF5
 
 | | Agonist bound human A2a adenosine receptor with D52N mutation at 2.60 A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 6-(2,2-diphenylethylamino)-9-[(2R,3R,4S,5S)-5-(ethylcarbamoyl)-3,4-dihydroxy-oxolan-2-yl]-N-[2-[(1-pyridin-2-ylpiperidin-4-yl)carbamoylamino]ethyl]purine-2-carboxamide, Human A2a adenosine receptor T4L chimera | | Authors: | White, K.L, Eddy, M.T, Gao, Z, Han, G.W, Hanson, M.A, Lian, T, Deary, A, Patel, N, Jacobson, K.A, Katritch, V, Stevens, R.C. | | Deposit date: | 2017-07-11 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Connection between Activation Microswitch and Allosteric Sodium Site in GPCR Signaling.
Structure, 26, 2018
|
|
8THK
 
 | |
149L
 
 | |
170L
 
 | | PROTEIN FLEXIBILITY AND ADAPTABILITY SEEN IN 25 CRYSTAL FORMS OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Zhang, X.-J, Weaver, L.H, Wozniak, A, Matthews, B.W. | | Deposit date: | 1995-03-24 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein flexibility and adaptability seen in 25 crystal forms of T4 lysozyme.
J.Mol.Biol., 250, 1995
|
|
1JQU
 
 | |
4XES
 
 | | Structure of active-like neurotensin receptor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krumm, B.E, White, J.F, Shah, P, Grisshammer, R. | | Deposit date: | 2014-12-24 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural prerequisites for G-protein activation by the neurotensin receptor.
Nat Commun, 6, 2015
|
|
