6CCD
 
 | |
2H34
 
 | | Apoenzyme crystal structure of the tuberculosis serine/threonine kinase, PknE | | Descriptor: | BROMIDE ION, SODIUM ION, Serine/threonine-protein kinase pknE | | Authors: | Gay, L.M, Ng, H.L, Alber, T. | | Deposit date: | 2006-05-22 | | Release date: | 2006-07-18 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Conserved Dimer and Global Conformational Changes in the Structure of apo-PknE Ser/Thr Protein Kinase from Mycobacterium tuberculosis.
J.Mol.Biol., 360, 2006
|
|
1T6O
 
 | | Nucleocapsid-binding domain of the measles virus P protein (amino acids 457-507) in complex with amino acids 486-505 of the measles virus N protein | | Descriptor: | linker, phosphoprotein | | Authors: | Kingston, R.L, Hamel, D.J, Gay, L.S, Dahlquist, F.W, Matthews, B.W. | | Deposit date: | 2004-05-06 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the attachment of a paramyxoviral polymerase to its template.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1P5C
 
 | |
1P56
 
 | |
2Y8U
 
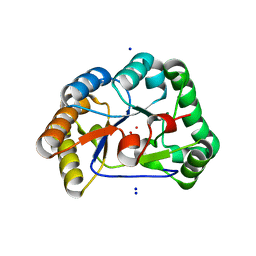 | | A. nidulans chitin deacetylase | | Descriptor: | CHITIN DEACETYLASE, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Penman, G, Gay, L.M, van Aalten, D.M.F. | | Deposit date: | 2011-02-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure and function of a broad-specificity chitin deacetylase from Aspergillus nidulans FGSC A4.
Sci Rep, 7, 2017
|
|
3B2X
 
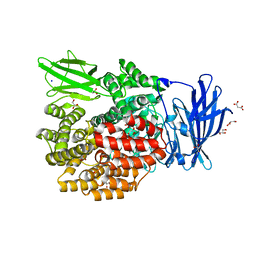 | | Crystal Structure of E. coli Aminopeptidase N in complex with Lysine | | Descriptor: | Aminopeptidase N, GLYCEROL, LYSINE, ... | | Authors: | Addlagatta, A, Gay, L, Matthews, B.W. | | Deposit date: | 2007-10-19 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the unusual specificity of Escherichia coli aminopeptidase N.
Biochemistry, 47, 2008
|
|
3BBZ
 
 | | Structure of the nucleocapsid-binding domain from the mumps virus phosphoprotein | | Descriptor: | BROMIDE ION, FORMIC ACID, P protein | | Authors: | Kingston, R.L, Gay, L.S, Baase, W.S, Matthews, B.W. | | Deposit date: | 2007-11-11 | | Release date: | 2008-05-27 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the nucleocapsid-binding domain from the mumps virus polymerase; an example of protein folding induced by crystallization
J.Mol.Biol., 379, 2008
|
|
2HPT
 
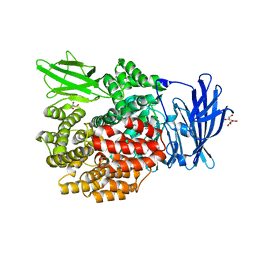 | | Crystal Structure of E. coli PepN (Aminopeptidase N)in complex with Bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Aminopeptidase N, GLYCEROL, ... | | Authors: | Addlagatta, A, Matthews, B.W, Gay, L. | | Deposit date: | 2006-07-17 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of aminopeptidase N from Escherichia coli suggests a compartmentalized, gated active site.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HPO
 
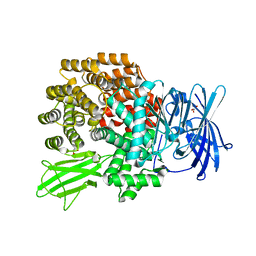 | | Structure of Aminopeptidase N from E. coli Suggests a Compartmentalized, Gated Active Site | | Descriptor: | Aminopeptidase N, GLYCEROL, ZINC ION | | Authors: | Addlagatta, A, Matthews, B.W, Gay, L. | | Deposit date: | 2006-07-17 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of aminopeptidase N from Escherichia coli suggests a compartmentalized, gated active site.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1OYU
 
 | |
3EOJ
 
 | | Fmo protein from Prosthecochloris Aestuarii 2K AT 1.3A Resolution | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, BACTERIOCHLOROPHYLL A, ... | | Authors: | Tronrud, D.E, Wen, J, Gay, L, Blankenship, R.E. | | Deposit date: | 2008-09-27 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structural basis for the difference in absorbance spectra for the FMO antenna protein from various green sulfur bacteria.
Photosynth.Res., 100, 2009
|
|
2Y8V
 
 | | Structure of chitinase, ChiC, from Aspergillus fumigatus. | | Descriptor: | CLASS III CHITINASE, PUTATIVE, SODIUM ION | | Authors: | Rush, C.L, Schuettelkopf, A.W, Gay, L.M, van Aalten, D.M.F. | | Deposit date: | 2011-02-10 | | Release date: | 2012-02-22 | | Last modified: | 2014-07-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of Chitinase, Chic, from Aspergillus Fumigatus.
To be Published
|
|
