1P5C
 
 | |
1P56
 
 | |
1OYU
 
 | |
2HPT
 
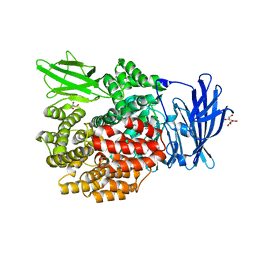 | | Crystal Structure of E. coli PepN (Aminopeptidase N)in complex with Bestatin | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Aminopeptidase N, GLYCEROL, ... | | Authors: | Addlagatta, A, Matthews, B.W, Gay, L. | | Deposit date: | 2006-07-17 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of aminopeptidase N from Escherichia coli suggests a compartmentalized, gated active site.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HPO
 
 | | Structure of Aminopeptidase N from E. coli Suggests a Compartmentalized, Gated Active Site | | Descriptor: | Aminopeptidase N, GLYCEROL, ZINC ION | | Authors: | Addlagatta, A, Matthews, B.W, Gay, L. | | Deposit date: | 2006-07-17 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of aminopeptidase N from Escherichia coli suggests a compartmentalized, gated active site.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3B2X
 
 | | Crystal Structure of E. coli Aminopeptidase N in complex with Lysine | | Descriptor: | Aminopeptidase N, GLYCEROL, LYSINE, ... | | Authors: | Addlagatta, A, Gay, L, Matthews, B.W. | | Deposit date: | 2007-10-19 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the unusual specificity of Escherichia coli aminopeptidase N.
Biochemistry, 47, 2008
|
|
3EOJ
 
 | | Fmo protein from Prosthecochloris Aestuarii 2K AT 1.3A Resolution | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, BACTERIOCHLOROPHYLL A, ... | | Authors: | Tronrud, D.E, Wen, J, Gay, L, Blankenship, R.E. | | Deposit date: | 2008-09-27 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structural basis for the difference in absorbance spectra for the FMO antenna protein from various green sulfur bacteria.
Photosynth.Res., 100, 2009
|
|
