5FL0
 
 | | Structure of a hydrolase with an inhibitor | | Descriptor: | (3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-2-(butylamino)-5-(hydroxymethyl)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d] [1,3]thiazole-6,7-diol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
6I6K
 
 | | Papaver somniferum O-methyltransferase 1 | | Descriptor: | (13aS)-3,10-dimethoxy-5,8,13,13a-tetrahydro-6H-isoquino[3,2-a]isoquinoline-2,9-diol, O-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Cabry, M.P, Offen, W.A, Winzer, T, Li, Y, Graham, I.A, Davies, G.J, Saleh, P. | | Deposit date: | 2018-11-15 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of Papaver somniferum O-Methyltransferase 1 Reveals Initiation of Noscapine Biosynthesis with Implications for Plant Natural Product Methylation
Acs Catalysis, 2019
|
|
5WJG
 
 | | Using sound pulses to solve the crystal harvesting bottleneck | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, L(+)-TARTARIC ACID, ... | | Authors: | Soares, A.S, Brennan, H.M, McCarthy, L, Leroy, L. | | Deposit date: | 2017-07-22 | | Release date: | 2017-08-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Using sound pulses to solve the crystal-harvesting bottleneck.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7BV9
 
 | | The NMR structure of the BEN domain from human NAC1 | | Descriptor: | Nucleus accumbens-associated protein 1 | | Authors: | Nagata, T, Kobayashi, N, Nakayama, N, Obayashi, E, Urano, T. | | Deposit date: | 2020-04-09 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Nucleus Accumbens-Associated Protein 1 Binds DNA Directly through the BEN Domain in a Sequence-Specific Manner.
Biomedicines, 8, 2020
|
|
6QGP
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-0769 | | Descriptor: | 1-cycloheptyl-3-[3-(cyclopentyloxy)-4-methoxyphenyl]-4,4-dimethyl-4,5-dihydro-1H-pyrazol-5-one, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Singh, A.K, Blaazer, A.R, Zara, L, de Esch, I.J.P, Leurs, R, Brown, D.G. | | Deposit date: | 2019-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-0769
To be published
|
|
7UJN
 
 | | Structure of Human SAMHD1 with Non-Hydrolysable dGTP Analog | | Descriptor: | 2'-deoxyguanosine-5'-O-(1-thiotriphosphate), Deoxynucleoside triphosphate triphosphohydrolase SAMHD1 | | Authors: | Huynh, K.W, Ammirati, M, Han, S. | | Deposit date: | 2022-03-31 | | Release date: | 2022-07-20 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Phosphorylation of SAMHD1 Thr592 increases C-terminal domain dynamics, tetramer dissociation and ssDNA binding kinetics.
Nucleic Acids Res., 50, 2022
|
|
8TQ7
 
 | | Crystal structure of Fab.34.2.12 in complex with MHC-I (H2-Dd) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Fab 34.2.12 Light Chain, ... | | Authors: | Jiang, J, Boyd, L.F, Natarajan, K, Margulies, D.H. | | Deposit date: | 2023-08-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Experimental Structures of Antibody/MHC-I Complexes Reveal Details of Epitopes Overlooked by Computational Prediction.
J Immunol., 212, 2024
|
|
7C6F
 
 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in an open state | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
7C6Y
 
 | | Crystal structure of beta-glycosides-binding protein (W41A) of ABC transporter in an open state (Form II) | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, CHLORIDE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-22 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
6UAY
 
 | | Crystal structure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) | | Descriptor: | GLYCOSIDE HYDROLASE | | Authors: | Costa, P.A.C.R, Santos, C.R, Domingues, M.N, Lima, E.A, Mandelli, F, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
2QKB
 
 | | Human RNase H catalytic domain mutant D210N in complex with 20-mer RNA/DNA hybrid | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*DGP*DGP*DAP*DAP*DTP*DCP*DAP*DGP*DGP*DTP*DGP*DTP*DCP*DGP*DCP*DAP*DCP*DTP*DCP*DT)-3', 5'-R(*GP*GP*AP*GP*UP*GP*CP*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*CP*C)-3'), ... | | Authors: | Nowotny, M, Gaidamakov, S.A, Ghirlando, R, Cerritelli, S.M, Crouch, R.J, Yang, W. | | Deposit date: | 2007-07-10 | | Release date: | 2007-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Human RNase H1 Complexed with an RNA/DNA Hybrid: Insight into HIV Reverse Transcription
Mol.Cell, 28, 2007
|
|
1KLJ
 
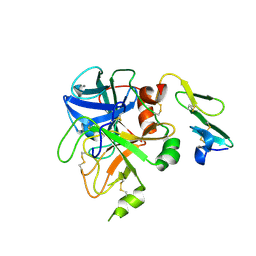 | | Crystal structure of uninhibited factor VIIa | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, factor VIIa | | Authors: | Sichler, K, Banner, D, D'Arcy, A, Hopfner, K.P, Huber, R, Bode, W, Kresse, G.B, Kopetzki, E, Brandstetter, H. | | Deposit date: | 2001-12-12 | | Release date: | 2002-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structures of uninhibited factor VIIa link its cofactor and substrate-assisted activation to specific interactions.
J.Mol.Biol., 322, 2002
|
|
5UAI
 
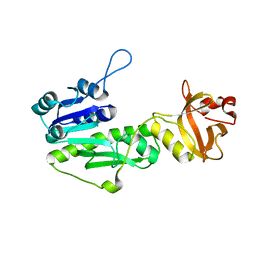 | |
4Z7B
 
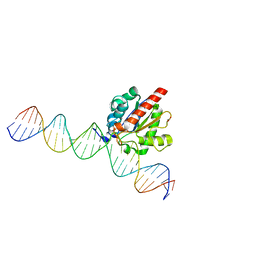 | | Structure of the enzyme-product complex resulting from TDG action on a GfC mismatch | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DNA (28-MER), ... | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2015-04-07 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA.
Nucleic Acids Res., 43, 2015
|
|
5GNS
 
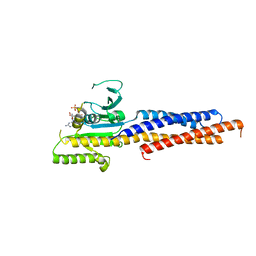 | | Structures of human Mitofusin 1 provide insight into mitochondrial tethering | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Mitofusin-1 | | Authors: | Qi, Y, Yan, L, Yu, C, Guo, X, Zhou, X, Hu, X, Huang, X, Rao, Z, Lou, Z, Hu, J. | | Deposit date: | 2016-07-24 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Structures of human Mitofusin 1 provide insight into mitochondrial tethering
To Be Published
|
|
8P0T
 
 | | CryoEM structure of 20S Trichomonas vaginalis proteasome in complex with proteasome inhibitor CP-17 | | Descriptor: | Family T1, proteasome alpha subunit, threonine peptidase, ... | | Authors: | Silhan, J, Boura, E, Fajtova, P. | | Deposit date: | 2023-05-10 | | Release date: | 2024-05-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural elucidation of recombinant Trichomonas vaginalis 20S proteasome bound to covalent inhibitors.
Nat Commun, 15, 2024
|
|
4ZAR
 
 | | Crystal Structure of Proteinase K from Engyodontium albuminhibited by METHOXYSUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYL KETONE at 1.15 A resolution | | Descriptor: | CALCIUM ION, METHOXYSUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYL KETONE, bound form, ... | | Authors: | Sawaya, M.R, Cascio, D, Collazo, M, Bond, C, Cohen, A, DeNicola, A, Eden, K, Jain, K, Leung, C, Lubock, N, McCormick, J, Rosinski, J, Spiegelman, L, Athar, Y, Tibrewal, N, Winter, J, Solomon, S. | | Deposit date: | 2015-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of Proteinase K from Engyodontium album inhibited by METHOXYSUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYL KETONE at 1.15 A resolution
to be published
|
|
9CX9
 
 | | Structure of SH3 domain of Src in complex with beta-arrestin 1 | | Descriptor: | Antibody fragment Fab30, heavy chain, light chain, ... | | Authors: | Pakharukova, N, Bansia, H, Lefkowitz, R.J, des Georges, A. | | Deposit date: | 2024-07-31 | | Release date: | 2024-11-13 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Beta-arrestin 1 mediated Src activation via Src SH3 domain revealed by cryo-electron microscopy.
Biorxiv, 2024
|
|
8P08
 
 | | Crystal structure of human CLK1 in complex with Leucettinib-21 | | Descriptor: | (4~{Z})-4-(1,3-benzothiazol-6-ylmethylidene)-2-[[(2~{R})-1-methoxy-4-methyl-pentan-2-yl]amino]-1~{H}-imidazol-5-one, Dual specificity protein kinase CLK1 | | Authors: | Kraemer, A, Schroeder, M, Meijer, L, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chemical, Biochemical, Cellular, and Physiological Characterization of Leucettinib-21, a Down Syndrome and Alzheimer's Disease Drug Candidate.
J.Med.Chem., 66, 2023
|
|
9D0I
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with Se-cresomycin, mRNA, deacylated A-site tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.45A resolution | | Descriptor: | (4S,5aS,8S,8aR)-4-(2-methylpropyl)-N-[(1R,5Z,7R,8R,9R,10R,11S,12R)-10,11,12-trihydroxy-7-methyl-13-oxa-2-selenabicyclo[7.3.1]tridec-5-en-8-yl]octahydro-2H-oxepino[2,3-c]pyrrole-8-carboxamide (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Robinson, P.J, Benedetto, A.E, Yu, M, Tresco, B.I.C, See, D.N.Y, Jiang, T, Ramkissoon, A, Dunand, C.F, Svetlov, M.S, Lee, J, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2024-08-07 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Why Sulfur is Important in Lincosamide Antibiotics.
Chem, 11, 2025
|
|
8TRV
 
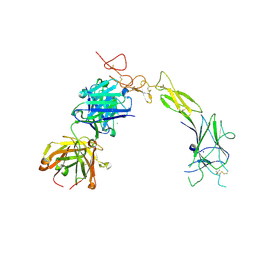 | | Structure of the EphA2 LBDCRD bound to FabS1C_C1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ephrin type-A receptor 2, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2024-07-17 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Synthetic antibodies targeting EphA2 induce diverse signaling-competent clusters with differential activation.
Protein Sci., 34, 2025
|
|
9D0H
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with C-cresomycin, mRNA, deacylated A-site tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.50A resolution | | Descriptor: | (4S,5aS,8S,8aR)-4-(2-methylpropyl)-N-[(1R,2R,3R,4Z,9R,10R,11R,12R)-10,11,12-trihydroxy-3-methyl-13-oxabicyclo[7.3.1]tridec-4-en-2-yl]octahydro-2H-oxepino[2,3-c]pyrrole-8-carboxamide, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Robinson, P.J, Benedetto, A.E, Yu, M, Tresco, B.I.C, See, D.N.Y, Jiang, T, Ramkissoon, A, Dunand, C.F, Svetlov, M.S, Lee, J, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2024-08-07 | | Release date: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Why Sulfur is Important in Lincosamide Antibiotics.
Chem, 11, 2025
|
|
5EDE
 
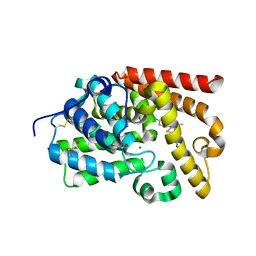 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c13c(cc(s1)C(NCC2OCCC2)=O)c(nn3c4ccc(cc4)Cl)C, micromolar IC50=0.217 | | Descriptor: | 1-(4-chlorophenyl)-3-methyl-~{N}-[[(2~{R})-oxolan-2-yl]methyl]thieno[2,3-c]pyrazole-5-carboxamide, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Joseph, C, Rudolph, M.G. | | Deposit date: | 2015-10-21 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
8OTR
 
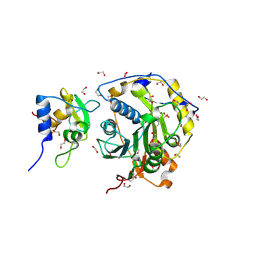 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with SAM analog BDH 33959089 | | Descriptor: | (2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-N-(1-methylpiperidin-4-yl)-3,4-bis(oxidanyl)oxolane-2-carboxamide, 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
8OW2
 
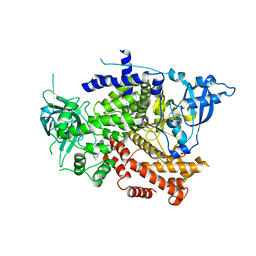 | | Crystal structure of the p110alpha catalytic subunit from homo sapiens in complex with activator 1938 | | Descriptor: | 1-[7-[[2-[[4-(4-ethylpiperazin-1-yl)phenyl]amino]pyridin-4-yl]amino]-2,3-dihydroindol-1-yl]ethanone, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gong, G.Q, Bellini, D, Vanhaesebroeck, B, Williams, R.L. | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
