8AH2
 
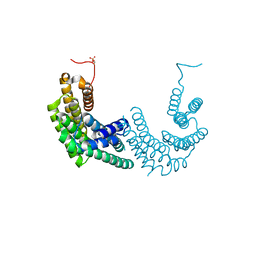 | | Crystal structure of human 14-3-3 zeta fused to the NPM1 peptide including phosphoserine-48 | | Descriptor: | 14-3-3 protein zeta/delta,Nucleophosmin | | Authors: | Boyko, K.M, Kapitonova, A.A, Tugaeva, K.V, Varfolomeeva, L.A, Sluchanko, N.N. | | Deposit date: | 2022-07-20 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the recognition by 14-3-3 proteins of a conditional binding site within the oligomerization domain of human nucleophosmin.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
6D4A
 
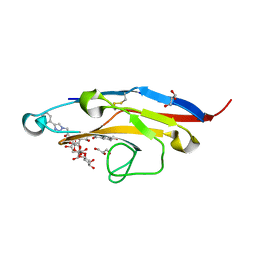 | | Cell Surface Receptor with Bound Ligand at 1.75-A Resolution | | Descriptor: | 2-aminoethyl 5-{[(4-cyclohexyl-1H-1,2,3-triazol-1-yl)acetyl]amino}-3,5,9-trideoxy-9-[(4-hydroxy-3,5-dimethylbenzene-1-carbonyl)amino]-D-glycero-alpha-D-galacto-non-2-ulopyranonosyl-(2->6)-beta-D-galactopyranosyl-(1->4)-beta-D-glucopyranoside, GLYCEROL, Myeloid cell surface antigen CD33 | | Authors: | Hermans, S.J, Miles, L.A, Parker, M.W. | | Deposit date: | 2018-04-17 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Small Molecule Binding to Alzheimer Risk Factor CD33 Promotes A beta Phagocytosis.
Iscience, 19, 2019
|
|
7B20
 
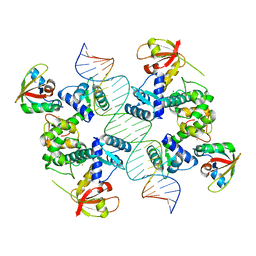 | |
8AHE
 
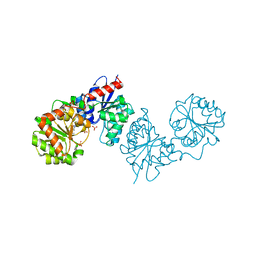 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | SULFATE ION, UDP-N-acetylglucosamine 2-epimerase, ~{N},5-dimethyl-1-phenyl-pyrazole-4-sulfonamide | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
8AHF
 
 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | (2~{R},4~{S})-4-[bis(fluoranyl)methoxy]-~{N}-methyl-1-(2~{H}-pyrazolo[4,3-b]pyridin-6-ylcarbonyl)pyrrolidine-2-carboxamide, SULFATE ION, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.271 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
6JOE
 
 | | Crystal structure of TrmD from Pseudomonas aeruginosa in complex with active-site inhibitor | | Descriptor: | PHOSPHATE ION, S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase, ... | | Authors: | Zhong, W, Pasunooti, K.K, Balamkundu, S, Wong, Y.W, Nah, Q, Liu, C.F, Lescar, J, Dedon, P.C. | | Deposit date: | 2019-03-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Thienopyrimidinone Derivatives That Inhibit Bacterial tRNA (Guanine37-N1)-Methyltransferase (TrmD) by Restructuring the Active Site with a Tyrosine-Flipping Mechanism.
J.Med.Chem., 62, 2019
|
|
7MF4
 
 | |
6JIS
 
 | | Crystal structure of the histidine racemase CntK in cobalt and nickel transporter system of staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, CESIUM ION, CHLORIDE ION, ... | | Authors: | Luo, S, Ju, Y, Zhou, H. | | Deposit date: | 2019-02-23 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of CntK, the cofactor-independent histidine racemase in staphylopine-mediated metal acquisition of Staphylococcus aureus.
Int.J.Biol.Macromol., 135, 2019
|
|
6DOD
 
 | |
7B1V
 
 | |
7RN8
 
 | | Crystal structure of caspase-3 with inhibitor Ac-VD(Orn)VD-CHO | | Descriptor: | Ac-VD(Orn)VD-CHO, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | McCue, W, Finzel, B.C. | | Deposit date: | 2021-07-29 | | Release date: | 2022-01-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure-Based Design and Biological Evaluation of Novel Caspase-2 Inhibitors Based on the Peptide AcVDVAD-CHO and the Caspase-2-Mediated Tau Cleavage Sequence YKPVD314.
Acs Pharmacol Transl Sci, 5, 2022
|
|
6JJ6
 
 | | BRD4 in complex with 500 | | Descriptor: | 1-methyl-6-(3-(4-methylpiperazine-1-carbonyl)benzyl)-1,2a1,5a,6-tetrahydro-2H-pyrido[3',2':6,7]azepino[4,3,2-cd]isoindol-2-one, Bromodomain-containing protein 4 | | Authors: | Xu, J, Chen, Y, Jiang, F, Zhu, J. | | Deposit date: | 2019-02-25 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | BRD4 in complex with ZZM1
To Be Published
|
|
6DOP
 
 | |
7RIX
 
 | | RNA polymerase II elongation complex with hairpin polyamide Py-Im 1, scaffold 2 | | Descriptor: | 3-({3-[(3-{[4-({4-[(4-{[4-({(2R)-2-amino-4-[(1-methyl-4-{[1-methyl-4-({1-methyl-4-[(1-methyl-1H-imidazole-2-carbonyl)amino]-1H-imidazole-2-carbonyl}amino)-1H-pyrrole-2-carbonyl]amino}-1H-pyrrole-2-carbonyl)amino]butanoyl}amino)-1-methyl-1H-imidazole-2-carbonyl]amino}-1-methyl-1H-pyrrole-2-carbonyl)amino]-1-methyl-1H-pyrrole-2-carbonyl}amino)-1-methyl-1H-pyrrole-2-carbonyl]amino}propyl)(methyl)amino]propyl}carbamoyl)benzoic acid, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ALR
 
 | | Crystal structure of TD1-gatorbulin1 complex | | Descriptor: | (2~{R})-2-oxidanyl-2-[(6~{S},9~{S},12~{S},15~{S},17~{S})-6,10,12,17-tetramethyl-3-methylidene-7-oxidanyl-2,5,8,11,14-pentakis(oxidanylidene)-13-oxa-1,4,7,10-tetrazabicyclo[13.3.0]octadecan-9-yl]ethanamide, Designed Ankyrin Repeat Protein (DARPIN) D1, GLYCEROL, ... | | Authors: | Oliva, M.A, Diaz, J.F. | | Deposit date: | 2020-10-07 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Gatorbulin-1, a distinct cyclodepsipeptide chemotype, targets a seventh tubulin pharmacological site.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RA0
 
 | | LC3A in complex with Fragment 2-10 | | Descriptor: | (5-ethyl-2-methyl-1H-indol-3-yl)acetic acid, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Rouge, L, Steffek, M, Helgason, E, Dueber, E, Mulvihill, M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A Multifaceted Hit-Finding Approach Reveals Novel LC3 Family Ligands.
Biochemistry, 62, 2023
|
|
7RFB
 
 | | Crystal structure of broadly neutralizing antibody mAb1198 in complex with Hepatitis C virus envelope glycoprotein E2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Flyak, A.I, Bjorkman, P.J. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Analysis of antibodies from HCV elite neutralizers identifies genetic determinants of broad neutralization.
Immunity, 55, 2022
|
|
7M8R
 
 | | Complex structure of Methane monooxygenase hydroxylase and regulatory subunit with fluorosubstituted tryptophans | | Descriptor: | 1,1,1-tris(fluoranyl)propan-2-one, 1,2-ETHANEDIOL, BENZOIC ACID, ... | | Authors: | Johns, J.C, Banerjee, R, Shi, K, Semonis, M.M, Aihara, H, Pomerantz, W.C.K, Lipscomb, J.D. | | Deposit date: | 2021-03-30 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Soluble Methane Monooxygenase Component Interactions Monitored by 19 F NMR.
Biochemistry, 60, 2021
|
|
7AEL
 
 | | alpha 1-antitrypsin (C232S) complexed with GSK716 | | Descriptor: | Alpha-1-antitrypsin, SULFATE ION, ~{N}-[(1~{S},2~{R})-1-(3-fluoranyl-2-methyl-phenyl)-1-oxidanyl-pentan-2-yl]-2-oxidanylidene-1,3-dihydroindole-4-carboxamide | | Authors: | Chung, C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-03-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Development of a small molecule that corrects misfolding and increases secretion of Z alpha 1 -antitrypsin.
Embo Mol Med, 13, 2021
|
|
8AO8
 
 | | Specific covalent inhibitor(9) of ERK2 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2~{R})-2-(3-methylimidazol-4-yl)piperidin-1-yl]propan-1-one, Mitogen-activated protein kinase 1, ... | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
7AT2
 
 | |
8AOB
 
 | | Specific covalent inhibitor(12) of ERK2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Mitogen-activated protein kinase 1, SULFATE ION, ... | | Authors: | Cleasby, A. | | Deposit date: | 2022-08-08 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | X-ray Screening of an Electrophilic Fragment Library and Application toward the Development of a Novel ERK 1/2 Covalent Inhibitor.
J.Med.Chem., 65, 2022
|
|
8SHK
 
 | |
6DP3
 
 | |
7RIW
 
 | | RNA polymerase II elongation complex scaffold 2, without polyamide | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Dervan, P.B, Wang, D. | | Deposit date: | 2021-07-20 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | RNA polymerase II trapped on a molecular treadmill: Structural basis of persistent transcriptional arrest by a minor groove DNA binder.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
