8WEA
 
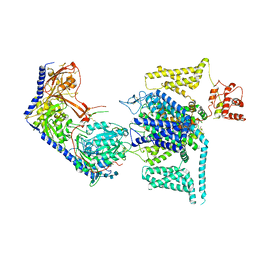 | | Human L-type voltage-gated calcium channel Cav1.2 (Class II) in the presence of pinaverium at 3.2 Angstrom resolution | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Fan, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
8WAY
 
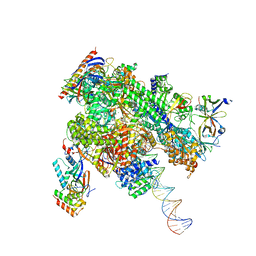 | | De novo transcribing complex 15 (TC15), the early elongation complex with Pol II positioned 15nt downstream of TSS | | Descriptor: | Alpha-amanitin, DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAW
 
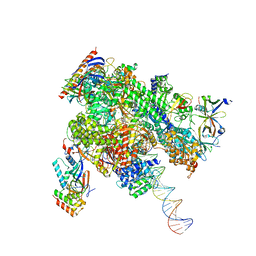 | | De novo transcribing complex 13 (TC13), the early elongation complex with Pol II positioned 13nt downstream of TSS | | Descriptor: | Alpha-amanitin, DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WE8
 
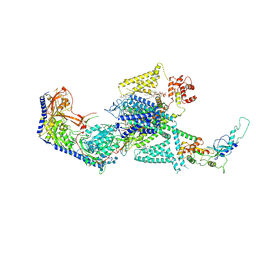 | | Human L-type voltage-gated calcium channel Cav1.2 in the presence of calciseptine, amlodipine and pinaverium at 2.9 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
8WE9
 
 | | Human L-type voltage-gated calcium channel Cav1.2 (Class I) in the presence of pinaverium at 3.0 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Fan, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
8WAT
 
 | | De novo transcribing complex 10 (TC10), the early elongation complex with Pol II positioned 10nt downstream of TSS | | Descriptor: | Alpha-amanitin, DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WB0
 
 | | De novo transcribing complex 17 (TC17), the early elongation complex with Pol II positioned 17nt downstream of TSS | | Descriptor: | Alpha-amanitin, DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WRM
 
 | | XBB.1.5 spike protein in complex with ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WAU
 
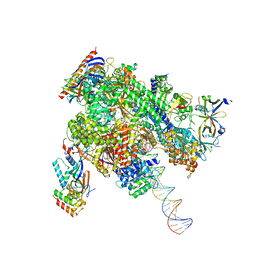 | | De novo transcribing complex 11 (TC11), the early elongation complex with Pol II positioned 11nt downstream of TSS | | Descriptor: | Alpha-amanitin, DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WE7
 
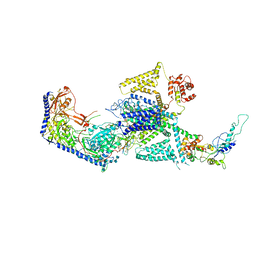 | | Human L-type voltage-gated calcium channel Cav1.2 in the presence of calciseptine at 3.2 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
8W77
 
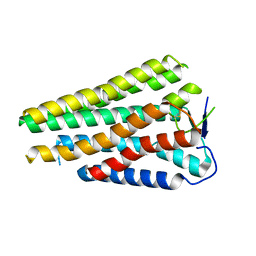 | | Human Consensus Olfactory Receptor OR52c in apo state, OR52c only | | Descriptor: | Human Consensus Olfactory Receptor OR52c in apo state, receptor only,Soluble cytochrome b562 | | Authors: | Choi, C.W, Bae, J, Choi, H.-J, Kim, J. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Understanding the molecular mechanisms of odorant binding and activation of the human OR52 family.
Nat Commun, 14, 2023
|
|
7V8I
 
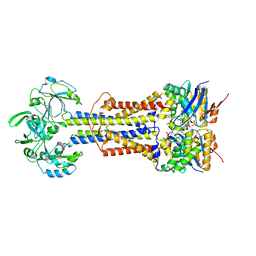 | | LolCD(E171Q)E with bound AMPPNP in nanodiscs | | Descriptor: | Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolC, Lipoprotein-releasing system transmembrane protein LolE, ... | | Authors: | Bei, W.W, Luo, Q.S, Shi, H.G, Zhang, X.Z, Huang, Y.H. | | Deposit date: | 2021-08-23 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of LolCDE reveal the molecular mechanism of bacterial lipoprotein sorting in Escherichia coli.
Plos Biol., 20, 2022
|
|
7VQF
 
 | | Phenol binding protein, MopR | | Descriptor: | ACETATE ION, PHENOL, Phenol sensing regulator, ... | | Authors: | Singh, J, Ray, S, Anand, R. | | Deposit date: | 2021-10-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Phenol sensing in nature is modulated via a conformational switch governed by dynamic allostery.
J.Biol.Chem., 298, 2022
|
|
7V8M
 
 | | LolCDE-apo in nanodiscs | | Descriptor: | Lipoprotein-releasing system ATP-binding protein LolD, Lipoprotein-releasing system transmembrane protein LolC, Lipoprotein-releasing system transmembrane protein LolE | | Authors: | Luo, Q.S, Bei, W.W, Shi, H.G, Zhang, X.Z, Huang, Y.H. | | Deposit date: | 2021-08-23 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of LolCDE reveal the molecular mechanism of bacterial lipoprotein sorting in Escherichia coli.
Plos Biol., 20, 2022
|
|
8WCC
 
 | | Cryo-EM structure of the CHA-bound mTAAR1 complex | | Descriptor: | CYCLOHEXYLAMMONIUM ION, Trace amine-associated receptor 1 | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WDT
 
 | | Crystal structure of the human adenosine A2A receptor in complex with photoresponsive ligand photoNECA(blue) | | Descriptor: | (2S,3S,4R,5R)-5-(6-amino-2-((E)-phenyldiazenyl)-9H-purin-9-yl)-N-ethyl-3,4-dihydroxytetrahydrofuran-2-carboxamide, Adenosine receptor A2a, Antibody Fab fragment heavy chain, ... | | Authors: | Araya, T, Asada, H, Iwata, S, Im, D.H. | | Deposit date: | 2023-09-16 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Crystal structure reveals the binding mode and selectivity of a photoswitchable ligand for the adenosine A 2A receptor.
Biochem.Biophys.Res.Commun., 695, 2023
|
|
8WRJ
 
 | | glycosyltransferase UGT74AN3 | | Descriptor: | 5-[(1R,2S,4R,6R,7R,10S,11S,14S,16R)-14-hydroxy-7,11-dimethyl-3-oxapentacyclo[8.8.0.02,4.02,7.011,16]octadecan-6-yl]pyran-2-one, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Huang, W, Long, F. | | Deposit date: | 2023-10-15 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8WRK
 
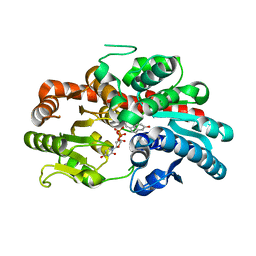 | | glycosyltransferase UGT74AN3 | | Descriptor: | Glycosyltransferase, PICEATANNOL, URIDINE-5'-DIPHOSPHATE | | Authors: | Huang, W, Long, F. | | Deposit date: | 2023-10-15 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8W8S
 
 | | Cryo-EM structure of the AA14-bound GPR101 complex | | Descriptor: | 1-(4-methylpyridin-2-yl)-3-[3-(trifluoromethyl)phenyl]thiourea, Probable G-protein coupled receptor 101 | | Authors: | Sun, J.P, Yu, X, Gao, N, Yang, F, Wang, J.Y, Yang, Z, Guan, Y, Wang, G.P. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
8VKP
 
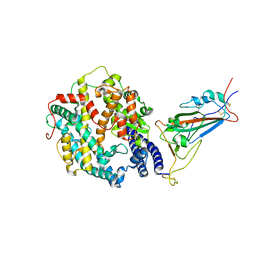 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
7VCE
 
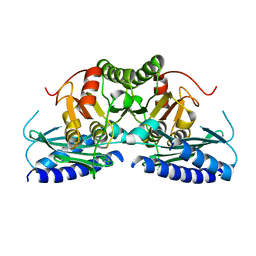 | | Structural studies of human inositol monophosphatase-1 inhibition by ebselen | | Descriptor: | Inositol monophosphatase 1 | | Authors: | Abuhammad, A, Laurieri, N, Rice, A, Lowe, E.D, McDonough, M.A, Singh, N, Churchill, G.C. | | Deposit date: | 2021-09-02 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical analysis of human inositol monophosphatase-1 inhibition by ebselen.
J.Biomol.Struct.Dyn., 2023
|
|
8VKO
 
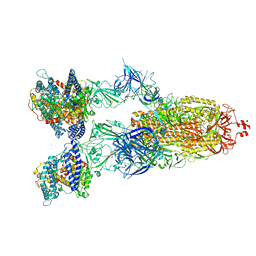 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
7VOZ
 
 | |
7VA8
 
 | | Crystal structure of MiCGT | | Descriptor: | UDP-glycosyltransferase 13, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Zhong, L, Zhang, Z.M. | | Deposit date: | 2021-08-27 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85003233 Å) | | Cite: | Directed Evolution of a Plant Glycosyltransferase for Chemo- and Regioselective Glycosylation of Pharmaceutically Significant Flavonoids
Acs Catalysis, 11, 2021
|
|
7VIB
 
 | | Crystal structure of human ACE2 and GX/P2V RBD | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein, ZINC ION | | Authors: | Guo, Y, Cao, W, Jia, N, Wang, W, Yuan, S, Wang, Y. | | Deposit date: | 2021-09-26 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of human ACE2 and GX/P2V RBD
To Be Published
|
|
