3KXX
 
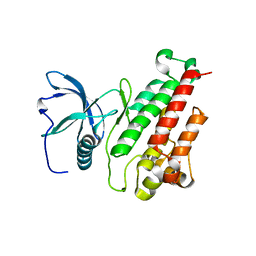 | | Structure of the mutant Fibroblast Growth Factor receptor 1 | | Descriptor: | Basic fibroblast growth factor receptor 1 | | Authors: | Bae, J.H, Boggon, T.J, Tome, F, Mandiyan, V, Lax, I, Schlessinger, J. | | Deposit date: | 2009-12-04 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Asymmetric receptor contact is required for tyrosine autophosphorylation of fibroblast growth factor receptor in living cells.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KY2
 
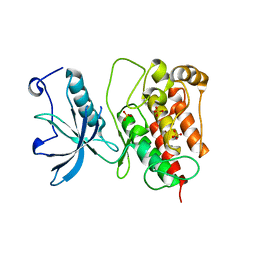 | | Crystal structure of Fibroblast Growth Factor Receptor 1 kinase domain | | Descriptor: | Basic fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Bae, J.H, Boggon, T.J, Tome, F, Mandiyan, V, Lax, I, Schlessinger, J. | | Deposit date: | 2009-12-04 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Asymmetric receptor contact is required for tyrosine autophosphorylation of fibroblast growth factor receptor in living cells.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3GQI
 
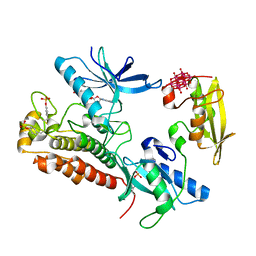 | | Crystal Structure of activated receptor tyrosine kinase in complex with substrates | | Descriptor: | Basic fibroblast growth factor receptor 1, DECAVANADATE, MAGNESIUM ION, ... | | Authors: | Bae, J.H, Lew, E.D, Yuzawa, S, Tome, F, Lax, I, Schlessinger, J. | | Deposit date: | 2009-03-24 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The selectivity of receptor tyrosine kinase signaling is controlled by a secondary SH2 domain binding site.
Cell(Cambridge,Mass.), 138, 2009
|
|
3GQL
 
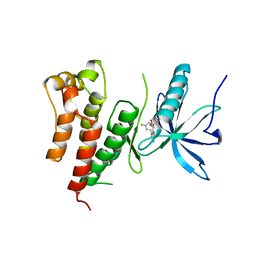 | | Crystal Structure of activated receptor tyrosine kinase in complex with substrates | | Descriptor: | (E)-[4-(3,5-difluorophenyl)-3H-pyrrolo[2,3-b]pyridin-3-ylidene](3-methoxyphenyl)methanol, Basic fibroblast growth factor receptor 1 | | Authors: | Bae, J.H, Lew, E.D, Yuzawa, S, Tome, F, Lax, I, Schlessinger, J. | | Deposit date: | 2009-03-24 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The selectivity of receptor tyrosine kinase signaling is controlled by a secondary SH2 domain binding site.
Cell(Cambridge,Mass.), 138, 2009
|
|
3JS2
 
 | | Crystal structure of minimal kinase domain of fibroblast growth factor receptor 1 in complex with 5-(2-thienyl)nicotinic acid | | Descriptor: | 5-(2-thienyl)nicotinic acid, Basic fibroblast growth factor receptor 1, PHOSPHATE ION | | Authors: | Bae, J.H, Ravindranathan, K.P, Mandiyan, V, Ekkati, A.R, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2009-09-09 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of novel fibroblast growth factor receptor 1 kinase inhibitors by structure-based virtual screening
J.Med.Chem., 53, 2010
|
|
1RRX
 
 | | Crystallographic Evidence for Isomeric Chromophores in 3-Fluorotyrosyl-Green Fluorescent Protein | | Descriptor: | SIGF1-GFP fusion protein | | Authors: | Bae, J.H, Paramita Pal, P, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-12-09 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Evidence for Isomeric Chromophores in 3-Fluorotyrosyl-Green Fluorescent Protein.
Chembiochem, 5, 2004
|
|
5Z6V
 
 | |
5Y8R
 
 | | ZsYellow at pH 3.5 | | Descriptor: | GFP-like fluorescent chromoprotein FP538 | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-21 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221.
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5Y8Q
 
 | | ZsYellow at pH 8.0 | | Descriptor: | GFP-like fluorescent chromoprotein FP538 | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-21 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221.
Biochem. Biophys. Res. Commun., 493, 2017
|
|
5Y4J
 
 | |
5Y4I
 
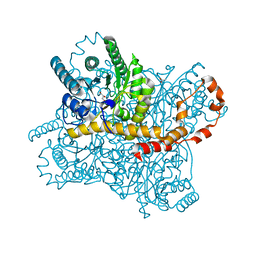 | | Crystal structure of glucose isomerase in complex with glycerol in one metal binding mode | | Descriptor: | ACETATE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Bae, J.E, Kim, I.J, Nam, K.H. | | Deposit date: | 2017-08-03 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of glucose isomerase in complex with xylitol inhibitor in one metal binding mode
Biochem. Biophys. Res. Commun., 493, 2017
|
|
8W77
 
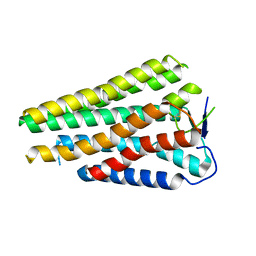 | | Human Consensus Olfactory Receptor OR52c in apo state, OR52c only | | Descriptor: | Human Consensus Olfactory Receptor OR52c in apo state, receptor only,Soluble cytochrome b562 | | Authors: | Choi, C.W, Bae, J, Choi, H.-J, Kim, J. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Understanding the molecular mechanisms of odorant binding and activation of the human OR52 family.
Nat Commun, 14, 2023
|
|
8GN5
 
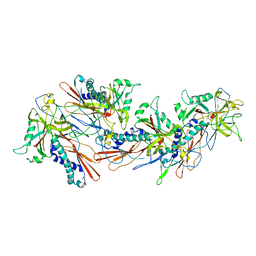 | | Designed pH-responsive P22 VLP | | Descriptor: | Major capsid protein | | Authors: | Kim, K.J, Kim, G, Bae, J.H, Song, J.J, Kim, H.S. | | Deposit date: | 2022-08-23 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | A pH-Responsive Virus-Like Particle as a Protein Cage for a Targeted Delivery.
Adv Healthc Mater, 13, 2024
|
|
7CM4
 
 | | Crystal Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody CT-P59 | | Descriptor: | 1,2-ETHANEDIOL, IgG heavy chain, IgG light chain, ... | | Authors: | Kim, Y.G, Jeong, J.H, Bae, J.S, Lee, J. | | Deposit date: | 2020-07-24 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein.
Nat Commun, 12, 2021
|
|
8HTI
 
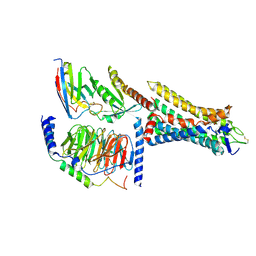 | | Human Consensus Olfactory Receptor OR52c in Complex with Octanoic Acid (OCA) and G Protein | | Descriptor: | Consensus Olfactory Receptor OR52c, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Choi, C.W, Bae, J, Choi, H.-J, Kim, J. | | Deposit date: | 2022-12-21 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Understanding the molecular mechanisms of odorant binding and activation of the human OR52 family.
Nat Commun, 14, 2023
|
|
8J46
 
 | | Human Consensus Olfactory Receptor OR52c in apo state, OR52c-bRIL | | Descriptor: | Olfactory receptor OR52c,Soluble cytochrome b562 | | Authors: | Choi, C.W, Bae, J, Choi, H.-J, Kim, J. | | Deposit date: | 2023-04-19 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Understanding the molecular mechanisms of odorant binding and activation of the human OR52 family.
Nat Commun, 14, 2023
|
|
3B2T
 
 | | Structure of phosphotransferase | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(methyl)phosphoryl]oxy}phosphoryl]adenosine, Fibroblast growth factor receptor 2, PHOSPHATE ION | | Authors: | Lew, E.D, Bae, J.H, Rohmann, E, Wollnik, B, Schlessinger, J. | | Deposit date: | 2007-10-19 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for reduced FGFR2 activity in LADD syndrome: Implications for FGFR autoinhibition and activation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3QF2
 
 | | Crystal structure of NALP3 PYD | | Descriptor: | NACHT, LRR and PYD domains-containing protein 3 | | Authors: | Park, H.H, Bae, J.Y. | | Deposit date: | 2011-01-21 | | Release date: | 2011-08-31 | | Last modified: | 2011-11-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of NALP3 Protein Pyrin Domain (PYD) and Its Implications in Inflammasome Assembly
J.Biol.Chem., 286, 2011
|
|
1OXD
 
 | | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins | | Descriptor: | cyan fluorescent protein cfp | | Authors: | Hyun Bae, J, Rubini, M, Jung, G, Wiegand, G, Seifert, M.H, Azim, M.K, Kim, J.S, Zumbusch, A, Holak, T.A, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-04-02 | | Release date: | 2003-12-02 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins
J.Mol.Biol., 328, 2003
|
|
1OXE
 
 | | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins | | Descriptor: | cyan fluorescent protein cfp | | Authors: | Hyun Bae, J, Rubini, M, Jung, G, Wiegand, G, Seifert, M.H, Azim, M.K, Kim, J.S, Zumbusch, A, Holak, T.A, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-04-02 | | Release date: | 2003-12-02 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins
J.Mol.Biol., 328, 2003
|
|
1OXF
 
 | | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins | | Descriptor: | cyan fluorescent protein cfp | | Authors: | Hyun Bae, J, Rubini, M, Jung, G, Wiegand, G, Seifert, M.H, Azim, M.K, Kim, J.S, Zumbusch, A, Holak, T.A, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-04-02 | | Release date: | 2003-12-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins
J.Mol.Biol., 328, 2003
|
|
2Q6P
 
 | | The Chemical Control of Protein Folding: Engineering a Superfolder Green Fluorescent Protein | | Descriptor: | Green fluorescent protein mutant 3 | | Authors: | Steiner, T, Hess, P, Bae, J.H, Wiltschi, B, Moroder, L, Budisa, N. | | Deposit date: | 2007-06-05 | | Release date: | 2007-06-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthetic biology of proteins: tuning GFPs folding and stability with fluoroproline.
Plos One, 3, 2008
|
|
1RM9
 
 | | Probing the Role of Tryptophans in Aequorea Victoria Green Fluorescent Proteins with an Expanded Genetic Code | | Descriptor: | avermectin-sensitive chloride channel GluCl beta/cyan fluorescent protein fusion | | Authors: | Budisa, N, Pal, P.P, Alefelder, S, Birle, P, Krywcun, T, Rubini, M, Wenger, W, Bae, J.H, Steiner, T. | | Deposit date: | 2003-11-27 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Probing the role of tryptophans in Aequorea victoria green fluorescent proteins with an expanded genetic code
Biol.Chem., 385, 2004
|
|
1RMP
 
 | | Probing the Role of Tryptophans in Aequorea Victoria Green Fluorescent Proteins with an Expanded Genetic Code | | Descriptor: | SIGF1-GFP fusion protein | | Authors: | Budisa, N, Pal, P.P, Alefelder, S, Birle, P, Krywcun, T, Rubini, M, Wenger, W, Bae, J.H, Steiner, T. | | Deposit date: | 2003-11-28 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the role of tryptophans in Aequorea victoria green fluorescent proteins with an expanded genetic code
Biol.Chem., 385, 2004
|
|
1RMO
 
 | | Probing the Role of Tryptophans in Aequorea Victoria Green Fluorescent Proteins with an Expanded Genetic Code | | Descriptor: | wunen-nonfunctional GFP fusion protein | | Authors: | Budisa, N, Pal, P.P, Alefelder, S, Birle, P, Krywcun, T, Rubini, M, Wenger, W, Bae, J.H, Steiner, T. | | Deposit date: | 2003-11-28 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the role of tryptophans in Aequorea victoria green fluorescent proteins with an expanded genetic code
Biol.Chem., 385, 2004
|
|
