4K6U
 
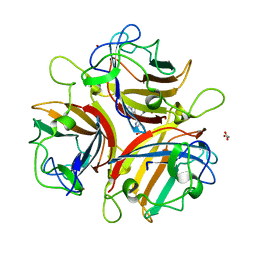 | | Crystal structure of Ad37 fiber knob in complex with trivalent sialic acid inhibitor ME0386 | | Descriptor: | 3,3',3''-[nitrilotris(methanediyl-1H-1,2,3-triazole-4,1-diyl)]tripropan-1-ol, ACETATE ION, Fiber protein, ... | | Authors: | Stehle, T, Bauer, J. | | Deposit date: | 2013-04-16 | | Release date: | 2014-10-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Triazole linker-based trivalent sialic acid inhibitors of adenovirus type 37 infection of human corneal epithelial cells.
Org.Biomol.Chem., 13, 2015
|
|
4KN1
 
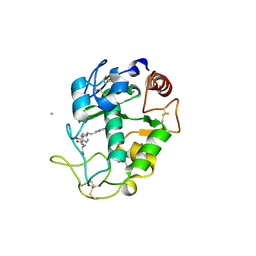 | |
5MO4
 
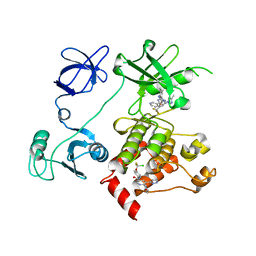 | |
2JHO
 
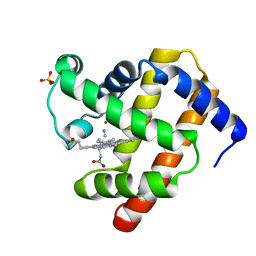 | | Cyanomet Sperm Whale Myoglobin at 1.4A resolution | | Descriptor: | CYANIDE ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Arcovito, A, Benfatto, M, Cianci, M, Hasnain, S.S, Nienhaus, K, Nienhaus, G.U, Savino, C, Strange, R.W, Vallone, B, Della Longa, S. | | Deposit date: | 2007-02-23 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray structure analysis of a metalloprotein with enhanced active-site resolution using in situ x-ray absorption near edge structure spectroscopy.
Proc. Natl. Acad. Sci. U.S.A., 104, 2007
|
|
2H9B
 
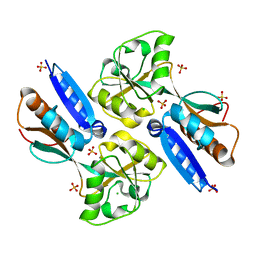 | | Crystal structure of the effector binding domain of a BenM variant (BenM R156H/T157S) | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator benM, SULFATE ION | | Authors: | Ezezika, O.C, Craven, S.H, Neidle, E.L, Momany, C. | | Deposit date: | 2006-06-09 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inducer responses of BenM, a LysR-type transcriptional regulator from Acinetobacter baylyi ADP1.
Mol.Microbiol., 72, 2009
|
|
1ZUA
 
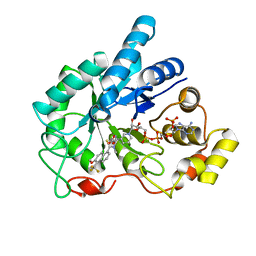 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Tolrestat | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TOLRESTAT | | Authors: | Gallego, O, Ruiz, F.X, Ardevol, A, Dominguez, M, Alvarez, R, de Lera, A.R, Rovira, C, Farres, J, Fita, I, Pares, X. | | Deposit date: | 2005-05-30 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural basis for the high all-trans-retinaldehyde reductase activity of the tumor marker AKR1B10.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2M8F
 
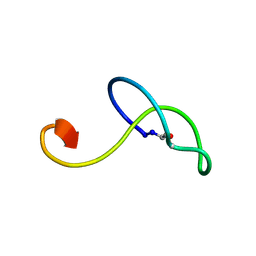 | | Structure of lasso peptide astexin3 | | Descriptor: | astexin3 | | Authors: | Maksimov, M.O, Link, A. | | Deposit date: | 2013-05-18 | | Release date: | 2013-07-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Discovery and characterization of an isopeptidase that linearizes lasso peptides.
J.Am.Chem.Soc., 135, 2013
|
|
8GK7
 
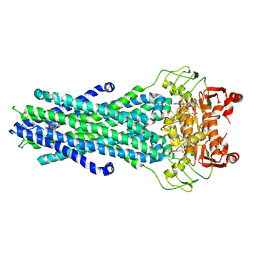 | | MsbA bound to cerastecin C | | Descriptor: | 2-[(4-butylbenzene-1-sulfonyl)amino]-5-[(3-{4-[(4-butylbenzene-1-sulfonyl)amino]-3-carboxyanilino}-3-oxopropyl)carbamoyl]benzoic acid, Lipid A export ATP-binding/permease protein MsbA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Chen, Y, Klein, D. | | Deposit date: | 2023-03-17 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cerastecins inhibit membrane lipooligosaccharide transport in drug-resistant Acinetobacter baumannii.
Nat Microbiol, 9, 2024
|
|
5L8C
 
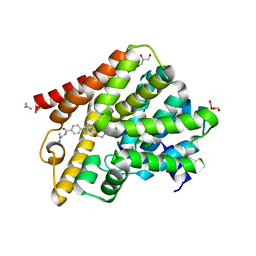 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-039 | | Descriptor: | 4-[5-[(4~{a}~{R},8~{a}~{S})-4-oxidanylidene-3-propan-2-yl-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-yl]-2-methoxy-phenyl]-~{N}-(2-azanyl-2-oxidanylidene-ethyl)benzamide, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Singh, A.K, Anthonyrajah, E.S, Brown, D.G. | | Deposit date: | 2016-06-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
5LAQ
 
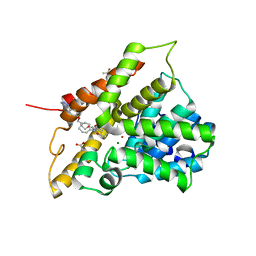 | | Crystal structure of human phosphodiesterase 4B catalytic domain with inhibitor NPD-001 | | Descriptor: | (4~{a}~{S},8~{a}~{R})-2-cycloheptyl-4-[4-methoxy-3-[4-[4-(1~{H}-1,2,3,4-tetrazol-5-yl)phenoxy]butoxy]phenyl]-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-one, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2016-06-14 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
5LBO
 
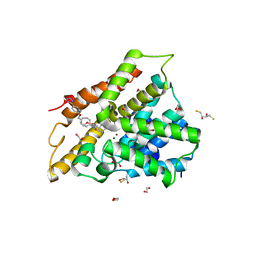 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-001 | | Descriptor: | (4~{a}~{S},8~{a}~{R})-2-cycloheptyl-4-[4-methoxy-3-[4-[4-(1~{H}-1,2,3,4-tetrazol-5-yl)phenoxy]butoxy]phenyl]-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-one, 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2016-06-16 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
8HS2
 
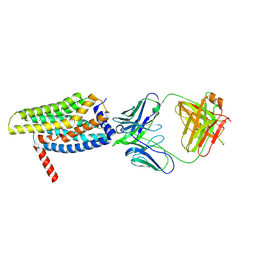 | | Orphan GPR20 in complex with Fab046 | | Descriptor: | Light chain of Fab046, Soluble cytochrome b562,G-protein coupled receptor 20, heavy chain of Fab046 | | Authors: | Lin, X, Jiang, S, Xu, F. | | Deposit date: | 2022-12-16 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
5KLG
 
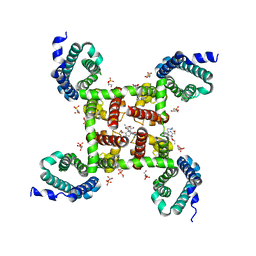 | | Structure of CavAb(W195Y) in complex with Br-dihydropyridine derivative UK-59811 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein, ... | | Authors: | Tang, L, Gamal EL-Din, T.M, Swanson, T.M, Pryde, D.C, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for inhibition of a voltage-gated Ca(2+) channel by Ca(2+) antagonist drugs.
Nature, 537, 2016
|
|
8HFM
 
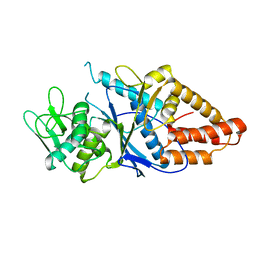 | | Crystal Structure of Mycobacterium smegmatis MshC | | Descriptor: | CALCIUM ION, L-cysteine:1D-myo-inositol 2-amino-2-deoxy-alpha-D-glucopyranoside ligase, ZINC ION | | Authors: | Pang, L, Weeks, S.D, Strelkov, S.V. | | Deposit date: | 2022-11-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural Basis of Cysteine Ligase MshC Inhibition by Cysteinyl-Sulfonamides.
Int J Mol Sci, 23, 2022
|
|
8HFO
 
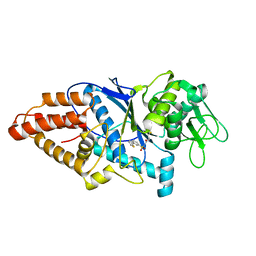 | | Crystal Structure of Mycobacterium smegmatis MshC in Complex with Compound 7d | | Descriptor: | CALCIUM ION, L-cysteine:1D-myo-inositol 2-amino-2-deoxy-alpha-D-glucopyranoside ligase, N-[(3M)-3-(thiophen-2-yl)benzene-1-sulfonyl]-L-cysteinamide, ... | | Authors: | Pang, L, Weeks, S.D, Strelkov, S.V. | | Deposit date: | 2022-11-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural Basis of Cysteine Ligase MshC Inhibition by Cysteinyl-Sulfonamides.
Int J Mol Sci, 23, 2022
|
|
8HFN
 
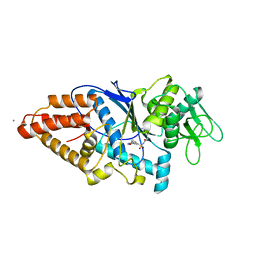 | | Crystal Structure of Mycobacterium smegmatis MshC in Complex with Compound 7b | | Descriptor: | CALCIUM ION, L-cysteine:1D-myo-inositol 2-amino-2-deoxy-alpha-D-glucopyranoside ligase, N-[(3M)-3-(6-methoxypyridin-3-yl)benzene-1-sulfonyl]-L-cysteinamide, ... | | Authors: | Pang, L, Weeks, S.D, Strelkov, S.V. | | Deposit date: | 2022-11-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Basis of Cysteine Ligase MshC Inhibition by Cysteinyl-Sulfonamides.
Int J Mol Sci, 23, 2022
|
|
8HGH
 
 | | Structure of 2:2 PAPP-A.STC2 complex | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Pappalysin-1, Stanniocalcin-2, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
8HGG
 
 | | Structure of 2:2 PAPP-A.ProMBP complex | | Descriptor: | Bone marrow proteoglycan, Pappalysin-1, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
8H3X
 
 | | Bacteroide Fragilis Toxin in complex with nanobody 282 | | Descriptor: | Fragilysin, ZINC ION, nanobody 282 | | Authors: | Wen, Y, Guo, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Screening and epitope characterization of diagnostic nanobody against total and activated Bacteroides fragilis toxin.
Front Immunol, 14, 2023
|
|
8H3Y
 
 | | Bacteroide Fragilis Toxin in complex with nanobody 327 | | Descriptor: | Fragilysin, Nanobody 327, ZINC ION | | Authors: | Wen, Y, Guo, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Screening and epitope characterization of diagnostic nanobody against total and activated Bacteroides fragilis toxin.
Front Immunol, 14, 2023
|
|
8HMV
 
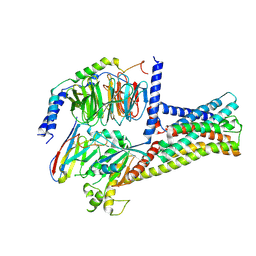 | | Structure of GPR21-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Wong, T.S, Gao, W. | | Deposit date: | 2022-12-05 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Cryo-EM structure of orphan G protein-coupled receptor GPR21.
MedComm (2020), 4, 2023
|
|
8HSC
 
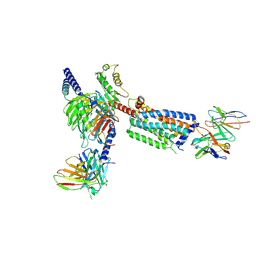 | | Gi bound Orphan GPR20 complex with Fab046 in ligand-free state | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Lin, X, Jiang, S, Xu, F. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
8HS3
 
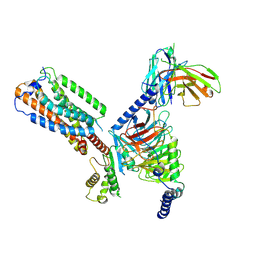 | | Gi bound orphan GPR20 in ligand-free state | | Descriptor: | Ggama, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Lin, X, Jiang, S, Xu, F. | | Deposit date: | 2022-12-16 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
8H3D
 
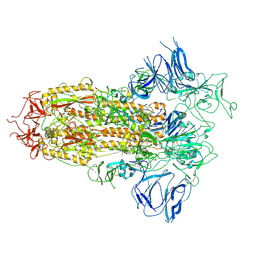 | | Structure of apo SARS-CoV-2 spike protein with one RBD up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Meng, F, Wang, Q, Xie, Y, Ni, X, Huang, N. | | Deposit date: | 2022-10-08 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | In Silico Discovery of Small Molecule Modulators Targeting the Achilles' Heel of SARS-CoV-2 Spike Protein.
Acs Cent.Sci., 9, 2023
|
|
8H3E
 
 | | Complex structure of a small molecule (SPC-14) bound SARS-CoV-2 spike protein, closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(6-nitro-2,3-dihydroindol-1-yl)-7-oxidanylidene-heptanoic acid, Spike glycoprotein,Fibritin | | Authors: | Meng, F, Wang, Q, Xie, Y, Ni, X, Huang, N. | | Deposit date: | 2022-10-08 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | In Silico Discovery of Small Molecule Modulators Targeting the Achilles' Heel of SARS-CoV-2 Spike Protein.
Acs Cent.Sci., 9, 2023
|
|
