8UT7
 
 | | CryoEM structure of A/Perth/16/2009 H3 in complex with polyclonal Fab from mice immunized with H3 stem nanoparticles-28 days post immunization | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H3D28 pFab HC Fv_polyA, ... | | Authors: | Huang, J, Han, J, Ward, A.B. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
8UT6
 
 | | CryoEM structure of A/Perth/16/2009 H3 in complex with polyclonal Fab from mice immunized with H3 stem nanoparticles-15 days post immunization | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H3D15 pFab HC Fv_polyA, ... | | Authors: | Huang, J, Han, J, Ward, A.B. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
8UT3
 
 | | CryoEM structure of A/Perth/16/2009 H3 in complex with flu HA central stem VH1-18 antibody UCA6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Huang, J, Han, J, Ward, A.B. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
8POT
 
 | | Ternary complex of E. coli leucyl-tRNA synthetase, tRNA(leu) and the benzoxaborole cmpd9 in the editing conformation | | Descriptor: | 1,2-ETHANEDIOL, Leucine--tRNA ligase, [(1R,3'S,5S,6R,8R)-3'-(aminomethyl)-8-(6-aminopurin-9-yl)-4'-bromanyl-7'-[3-[methyl-(phenylmethyl)amino]propoxy]spiro[2,4,7-trioxa-3$l^{4}-borabicyclo[3.3.0]octane-3,1'-3H-2,1$l^{4}-benzoxaborole]-6-yl]methyl dihydrogen phosphate, ... | | Authors: | Palencia, A, Hoffmann, G. | | Deposit date: | 2023-07-05 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Targeting a microbiota Wolbachian aminoacyl-tRNA synthetase to block its pathogenic host.
Sci Adv, 10, 2024
|
|
8POR
 
 | |
7UGH
 
 | |
4XJR
 
 | | The catalytic mechanism of human parainfluenza virus type 3 haemagglutinin-neuraminidase revealed | | Descriptor: | (6R)-2,6-anhydro-3,4,5-trideoxy-6-[(2S)-2,3-dihydroxypropanoyl]-3-fluoro-5-[(2-methylpropanoyl)amino]-4-triaza-1,2-dien -2-ium-1-yl-L-gulonic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dirr, L, El-Deeb, I, Guillon, P, Carroux, C, Chavas, L, von Itzstein, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-02-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The catalytic mechanism of human parainfluenza virus type 3 haemagglutinin-neuraminidase revealed.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5YZ1
 
 | | Crystal structure of human Archease | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Protein archease | | Authors: | Duan, S.Y, Li, J.X. | | Deposit date: | 2017-12-11 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of human archease, a key cofactor of tRNA splicing ligase complex.
Int.J.Biochem.Cell Biol., 122, 2020
|
|
7CBE
 
 | |
1F2S
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX FORMED BETWEEN BOVINE BETA-TRYPSIN AND MCTI-A, A TRYPSIN INHIBITOR OF SQUASH FAMILY AT 1.8 A RESOLUTION | | Descriptor: | CALCIUM ION, TRYPSIN, TRYPSIN INHIBITOR A | | Authors: | Zhu, Y, Huang, Q, Qian, M, Jia, Y, Tang, Y. | | Deposit date: | 2000-05-29 | | Release date: | 2000-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of the complex formed between bovine beta-trypsin and MCTI-A, a trypsin inhibitor of squash family, at 1.8-A resolution.
J.Protein Chem., 18, 1999
|
|
5JO3
 
 | | PDE5A for NaV1.7 | | Descriptor: | 1-(5-chloro-6-methoxypyridin-3-yl)-3-methyl-N-(methylsulfonyl)-1H-indazole-5-carboxamide, ZINC ION, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Storer, I, Chrencik, J. | | Deposit date: | 2016-05-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | PDE5A for NaV1.7
To be published
|
|
8VDS
 
 | | DNA Ligase 1 with nick DNA 3'rG:C | | Descriptor: | DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA ligase 1, DNA/RNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*T)-R(P*G)-D(P*GP*TP*CP*GP*GP*AP*C)-3') | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2023-12-17 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
8A55
 
 | |
8VZM
 
 | | DNA Ligase 1 captured with pre-step 3 ligation at the rA:T nicksite | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*GP*AP*C)-3'), ... | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2024-02-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
6Z52
 
 | | Crystal structure of CLK3 in complex with macrocycle ODS2003136 | | Descriptor: | 1,2-ETHANEDIOL, 11,15-dimethyl-6-(4-methylpiperazin-1-yl)-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, CHLORIDE ION, ... | | Authors: | Chaikuad, A, Benderitter, P, Hoflack, J, Denis, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of CLK3 in complex with macrocycle ODS2003136
To Be Published
|
|
8VDT
 
 | | DNA Ligase 1 with nick DNA 3'rA:T | | Descriptor: | DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*TP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA ligase 1, DNA/RNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*T)-R(P*A)-D(P*GP*TP*CP*GP*GP*AP*C)-3'), ... | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2023-12-17 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
8VZL
 
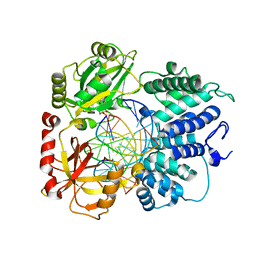 | | DNA Ligase 1 captured with pre-step 3 ligation at the rG:C nicksite | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*CP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*GP*AP*C)-3'), ... | | Authors: | KanalElamparithi, B, Gulkis, M, Caglayan, M. | | Deposit date: | 2024-02-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structures of LIG1 provide a mechanistic basis for understanding a lack of sugar discrimination against a ribonucleotide at the 3'-end of nick DNA.
J.Biol.Chem., 300, 2024
|
|
8ZWS
 
 | | Mtb. MazF-mt3 toxin in compleex with its antitoxin fragmant | | Descriptor: | Antitoxin MazE6, Endoribonuclease MazF6 | | Authors: | Xie, W, Jiang, P. | | Deposit date: | 2024-06-13 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Long Dynamic beta 1-beta 2 Loops in M. tb MazF Toxins Affect the Interaction Modes and Strengths of the Toxin-Antitoxin Pairs.
Int J Mol Sci, 25, 2024
|
|
4UCT
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 2-amino-6-methyl-5-(propan-2-yloxy)-3H-[1,2,4]triazolo[1,5-a]pyrimidin-8-ium, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
4UH5
 
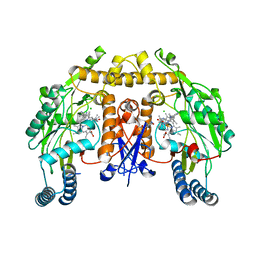 | | Structure of human nNOS R354A G357D mutant heme domain in complex with N1-(5-(2-(6-Amino-4-methylpyridin-2-yl)ethyl)pyridin-3-yl)-N1,N2- dimethylethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, CHLORIDE ION, N1-(5-(2-(6-Amino-4-methylpyridin-2-yl)ethyl)pyridin-3-yl)-N1,N2-dimethylethane-1,2-diamine, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-03-23 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | 2-Aminopyridines with a Truncated Side Chain to Improve Human Neuronal Nitric Oxide Synthase Inhibitory Potency and Selectivity.
J.Med.Chem., 58, 2015
|
|
8W1L
 
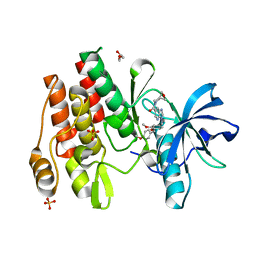 | |
1F4I
 
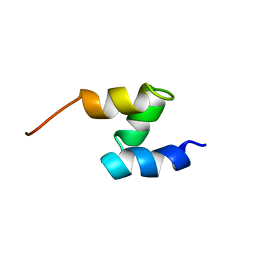 | | SOLUTION STRUCTURE OF THE HHR23A UBA(2) MUTANT P333E, DEFICIENT IN BINDING THE HIV-1 ACCESSORY PROTEIN VPR | | Descriptor: | UV EXCISION REPAIR PROTEIN PROTEIN RAD23 HOMOLOG A | | Authors: | Withers-Ward, E.S, Mueller, T.D, Chen, I.S, Feigon, J. | | Deposit date: | 2000-06-07 | | Release date: | 2000-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Biochemical and structural analysis of the interaction between the UBA(2) domain of the DNA repair protein HHR23A and HIV-1 Vpr.
Biochemistry, 39, 2000
|
|
5H7H
 
 | | Crystal structure of the BCL6 BTB domain in complex with F1324(10-13) | | Descriptor: | 1,2-ETHANEDIOL, B-cell lymphoma 6 protein, F1324 peptide residues 10-13 | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2016-11-18 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of high-affinity BCL6-binding peptide and its structure-activity relationship.
Biochem. Biophys. Res. Commun., 482, 2017
|
|
9FL0
 
 | | Discovery of a Series of Covalent, Cell Active Bfl-1 Inhibitors | | Descriptor: | Bcl-2-related protein A1, ~{N}-[(4-chlorophenyl)methyl]-~{N}-(4-fluorophenyl)prop-2-enamide | | Authors: | Hargreaves, D. | | Deposit date: | 2024-06-04 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-Based Optimization of a Series of Covalent, Cell Active Bfl-1 Inhibitors.
J.Med.Chem., 67, 2024
|
|
6FSN
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with UDP-glucose (conformation 1) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Roversi, P, Le Cornu, J.D, Hill, J, Alonzi, D.S, Zitzmann, N. | | Deposit date: | 2018-02-19 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Crystal polymorphism in fragment-based lead discovery of ligands of the catalytic domain of UGGT, the glycoprotein folding quality control checkpoint.
Front Mol Biosci, 2022
|
|
