6ZRR
 
 | |
6ZS2
 
 | | Crystal Structure of the bromodomain of human transcription activator BRG1 (SMARCA4) in complex with 2-(6-amino-5-(piperazin-1-yl)pyridazin-3-yl)phenol | | Descriptor: | 1,2-ETHANEDIOL, 2-(6-azanyl-5-piperazin-4-ium-1-yl-pyridazin-3-yl)phenol, Transcription activator BRG1 | | Authors: | Preuss, F, Joerger, A.C, Kraemer, A, Wanior, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-07-15 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Pan-SMARCA/PB1 Bromodomain Inhibitors and Their Role in Regulating Adipogenesis.
J.Med.Chem., 63, 2020
|
|
6ZS8
 
 | |
6ZPR
 
 | | Solution structure of MLKL executioner domain in complex with a covalent inhibitor | | Descriptor: | 7-(2-methoxyethoxymethyl)-1,3-dimethyl-purine-2,6-dione, Mixed lineage kinase domain-like protein,Mixed lineage kinase domain-like protein | | Authors: | Ruebbelke, M, Bauer, M, Hamilton, J, Binder, F, Nar, H, Zeeb, M. | | Deposit date: | 2020-07-09 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Locking mixed-lineage kinase domain-like protein in its auto-inhibited state prevents necroptosis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZTS
 
 | |
6ZTV
 
 | | Crystal Structure of catalase HPII from Escherichia coli (serendipitously crystallized) | | Descriptor: | 1,2-ETHANEDIOL, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Catalase HPII, ... | | Authors: | Grzechowiak, M, Sekula, B, Ruszkowski, M. | | Deposit date: | 2020-07-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Serendipitous crystallization of E. coli HPII catalase, a sequel to "the tale usually not told".
Acta Biochim.Pol., 68, 2021
|
|
6ZVO
 
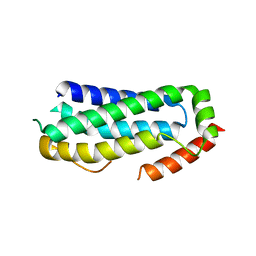 | | Crystal structure of unliganded MLKL executioner domain | | Descriptor: | BROMIDE ION, CHLORIDE ION, Mixed lineage kinase domain-like protein | | Authors: | Fiegen, D, Bauer, M, Nar, H. | | Deposit date: | 2020-07-27 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.371 Å) | | Cite: | Locking mixed-lineage kinase domain-like protein in its auto-inhibited state prevents necroptosis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7A4M
 
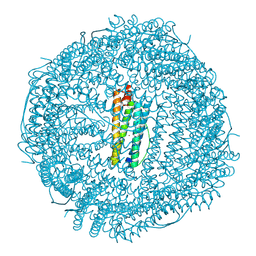 | | Cryo-EM structure of mouse heavy-chain apoferritin at 1.22 A | | Descriptor: | FE (III) ION, Ferritin heavy chain, ZINC ION | | Authors: | Nakane, T, Kotecha, A, Sente, A, Yamashita, K, McMullan, G, Masiulis, S, Brown, P.M.G.E, Grigoras, I.T, Malinauskaite, L, Malinauskas, T, Miehling, J, Yu, L, Karia, D, Pechnikova, E.V, de Jong, E, Keizer, J, Bischoff, M, McCormack, J, Tiemeijer, P, Hardwick, S.W, Chirgadze, D.Y, Murshudov, G, Aricescu, A.R, Scheres, S.H.W. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (1.22 Å) | | Cite: | Single-particle cryo-EM at atomic resolution.
Nature, 587, 2020
|
|
7A19
 
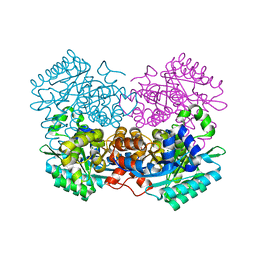 | |
7A2H
 
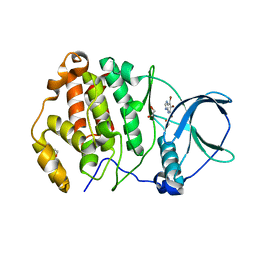 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the ATP-competitive inhibitor 5,6,7-tribromo-1H-imidazo[4,5-b]pyridine | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7-tris(bromanyl)-1~{H}-imidazo[4,5-b]pyridine, Casein kinase II subunit alpha' | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-18 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
7A41
 
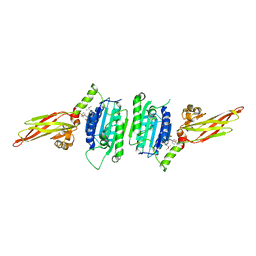 | | MALT1 in complex with a NVS-MALT1 chemical probe | | Descriptor: | DIMETHYL SULFOXIDE, Mucosa-associated lymphoid tissue lymphoma translocation protein 1, ~{N}-[(1~{S})-1-[4-[[2-chloranyl-7-[(1~{S})-1-methoxyethyl]pyrazolo[1,5-a]pyrimidin-6-yl]amino]phenyl]-2,2,2-tris(fluoranyl)ethyl]-~{N}-methyl-1,1-bis(oxidanylidene)thiane-4-carboxamide | | Authors: | Renatus, M. | | Deposit date: | 2020-08-19 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | MALT1 in complex with a NVS-MALT1 chemical probe
Not Published
|
|
7A4C
 
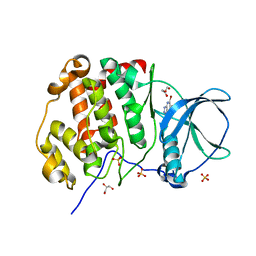 | | Crystal structure of human protein kinase CK2alpha (CSNK2A1 gene product) in complex with the ATP-competitive inhibitor 5,6,7-tribromo-1H-triazolo[4,5-b]pyridine | | Descriptor: | 5,6,7-tris(bromanyl)-1~{H}-[1,2,3]triazolo[4,5-b]pyridine, Casein kinase II subunit alpha, GLYCEROL, ... | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-19 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
5W4X
 
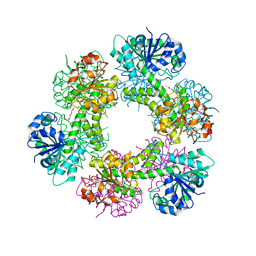 | | Truncated hUGDH | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, UDP-glucose 6-dehydrogenase | | Authors: | Sennett, N.C, Custer, G.S, Wood, Z.A. | | Deposit date: | 2017-06-13 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The entropic force generated by intrinsically disordered segments tunes protein function.
Nature, 563, 2018
|
|
6ZSO
 
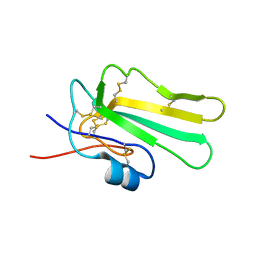 | | Solution structure of the water-soluble LU-domain of human Lypd6b protein | | Descriptor: | Ly6/PLAUR domain-containing protein 6B | | Authors: | Tsarev, A.V, Kulbatskii, D.S, Paramonov, A.S, Lyukmanova, E.N, Shenkarev, Z.O. | | Deposit date: | 2020-07-16 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural Diversity and Dynamics of Human Three-Finger Proteins Acting on Nicotinic Acetylcholine Receptors.
Int J Mol Sci, 21, 2020
|
|
6ZNV
 
 | | Protein polybromo-1 (PB1 BD2) Bound To DP28 | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]piperidin-4-ol, ACETATE ION, ... | | Authors: | Preuss, F, Mathea, S, Chatterjee, D, Wanior, M, Joerger, A.C, Knapp, S. | | Deposit date: | 2020-07-06 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Pan-SMARCA/PB1 Bromodomain Inhibitors and Their Role in Regulating Adipogenesis.
J.Med.Chem., 63, 2020
|
|
6ZTY
 
 | |
1VPO
 
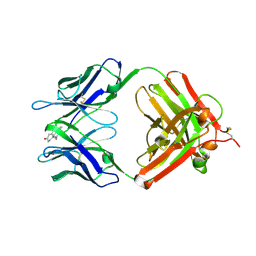 | | Crystal Structure Analysis of the Anti-testosterone Fab in Complex with Testosterone | | Descriptor: | TESTOSTERONE, anti-testosterone (heavy chain), anti-testosterone (light chain) | | Authors: | Valjakka, J.M, Hemminki, A, Niemi, S, Soderlund, H, Takkinen, K, Rouvinen, J. | | Deposit date: | 2004-11-15 | | Release date: | 2004-11-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of an in Vitro Affinity- and Specificity-matured Anti-testosterone
Fab in Complex with Testosterone. IMPROVED AFFINITY RESULTS FROM
SMALL STRUCTURAL CHANGES WITHIN THE VARIABLE DOMAINS
J.Biol.Chem., 277, 2002
|
|
6ZPG
 
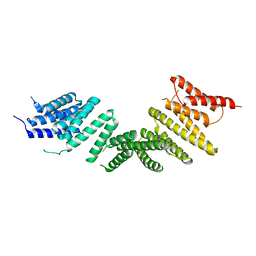 | | Kinesin binding protein (KBP) | | Descriptor: | KIF-binding protein | | Authors: | Atherton, J, Hummel, J.J.A, Olieric, N, Locke, J, Pena, A, Rosenfeld, S.S, Steinmetz, M.O, Hoogenraad, C.C, Moores, C.A. | | Deposit date: | 2020-07-08 | | Release date: | 2020-12-30 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The mechanism of kinesin inhibition by kinesin-binding protein.
Elife, 9, 2020
|
|
1VQW
 
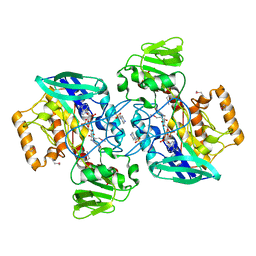 | | Crystal structure of a protein with similarity to flavin-containing monooxygenases and to mammalian dimethylalanine monooxygenases | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN WITH SIMILARITY TO FLAVIN-CONTAINING MONOOXYGENASES AND TO MAMMALIAN DIMETHYLALANINE MONOOXYGENASES | | Authors: | Eswaramoorthy, S, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-01-05 | | Release date: | 2005-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of action of a flavin-containing monooxygenase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6ZPJ
 
 | | Crystal structure of the unconventional kinetochore protein Leishmania mexicana KKT4 coiled coil domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, IMIDAZOLE, Leishmania mexicana KKT4 | | Authors: | Ludzia, P, Lowe, E.D, Marciano, G, Mohammed, S, Redfield, C, Akiyoshi, B. | | Deposit date: | 2020-07-08 | | Release date: | 2020-10-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of KKT4, an unconventional microtubule-binding kinetochore protein.
Structure, 29, 2021
|
|
6ZZ1
 
 | | Crystal structure of MLKL executioner domain in complex with a covalent inhibitor | | Descriptor: | 7-(2-methoxyethoxymethyl)-1,3-dimethyl-purine-2,6-dione, Mixed lineage kinase domain-like protein | | Authors: | Fiegen, D, Bauer, M, Nar, H. | | Deposit date: | 2020-08-03 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Locking mixed-lineage kinase domain-like protein in its auto-inhibited state prevents necroptosis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1VRO
 
 | | Selenium-Assisted Nucleic Acid Crystallography: Use of Phosphoroselenoates for MAD Phasing of a DNA Structure | | Descriptor: | 5'-D(*CP*(GMS)P*CP*GP*CP*G)-3', MAGNESIUM ION, SPERMINE | | Authors: | Wilds, C.J, Pattanayek, R, Pan, C, Wawrzak, Z, Egli, M. | | Deposit date: | 2005-04-14 | | Release date: | 2005-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Selenium-Assisted Nucleic Acid Crystallography: Use of Phosphoroselenoates for MAD Phasing of a DNA Structure
J.Am.Chem.Soc., 124, 2002
|
|
1VRP
 
 | | The 2.1 Structure of T. californica Creatine Kinase Complexed with the Transition-State Analogue Complex, ADP-Mg 2+ /NO3-/Creatine | | Descriptor: | (DIAMINOMETHYL-METHYL-AMINO)-ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, Creatine Kinase, ... | | Authors: | Lahiri, S.D, Wang, P.F, Babbitt, P.C, McLeish, M.J, Kenyon, G.L, Allen, K.N. | | Deposit date: | 2005-04-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1 A Structure of Torpedo californica Creatine Kinase Complexed with the ADP-Mg(2+)-NO3(-)-Creatine Transition-State Analogue Complex
Biochemistry, 41, 2002
|
|
1VRZ
 
 | | Helix turn helix motif | | Descriptor: | ACETATE ION, DE NOVO DESIGNED 21 RESIDUE PEPTIDE | | Authors: | Rudresh, Ramakumar, S, Ramagopal, U.A, Inai, Y, Sahal, D. | | Deposit date: | 2005-10-14 | | Release date: | 2005-11-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | De Novo Design and Characterization of a Helical Hairpin Eicosapeptide; Emergence of an Anion Receptor in the Linker Region.
Structure, 12, 2004
|
|
6ZN3
 
 | | Plasmodium facliparum glideosome trimeric sub-complex | | Descriptor: | Myosin A tail domain interacting protein, Myosin essential light chain ELC, Myosin-A | | Authors: | Pazicky, S, Loew, C. | | Deposit date: | 2020-07-06 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural role of essential light chains in the apicomplexan glideosome.
Commun Biol, 3, 2020
|
|
