1SQV
 
 | | Crystal Structure Analysis of Bovine Bc1 with UHDBT | | Descriptor: | 6-HYDROXY-5-UNDECYL-1,3-BENZOTHIAZOLE-4,7-DIONE, Cytochrome b, Cytochrome c1, ... | | Authors: | Esser, L, Quinn, B, Li, Y.F, Zhang, M, Elberry, M, Yu, L, Yu, C.A, Xia, D. | | Deposit date: | 2004-03-19 | | Release date: | 2005-09-06 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystallographic studies of quinol oxidation site inhibitors: a modified classification of inhibitors for the cytochrome bc(1) complex.
J.Mol.Biol., 341, 2004
|
|
2FFZ
 
 | |
1MIT
 
 | | RECOMBINANT CUCURBITA MAXIMA TRYPSIN INHIBITOR V (RCMTI-V) (NMR, MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | TRYPSIN INHIBITOR V | | Authors: | Cai, M, Gong, Y, Huang, Y, Liu, J, Prakash, O, Wen, L, Wen, J.J, Huang, J.-K, Krishnamoorthi, R. | | Deposit date: | 1995-10-26 | | Release date: | 1996-04-03 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of recombinant Cucurbita maxima trypsin inhibitor-V determined by NMR spectroscopy.
Biochemistry, 35, 1996
|
|
1MLY
 
 | | Crystal Structure of 7,8-Diaminopelargonic Acid Synthase in complex with the cis isomer of amiclenomycin | | Descriptor: | 7,8-diamino-pelargonic acid aminotransferase, CIS-AMICLENOMYCIN, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Sandmark, J, Mann, S, Marquet, A, Schneider, G. | | Deposit date: | 2002-09-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for the inhibition of the biosynthesis of biotin by the antibiotic amiclenomycin
J.Biol.Chem., 277, 2002
|
|
1MPQ
 
 | | MALTOPORIN TREHALOSE COMPLEX | | Descriptor: | MALTOPORIN, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Dutzler, R, Schirmer, T. | | Deposit date: | 1997-03-24 | | Release date: | 1998-03-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Channel specificity: structural basis for sugar discrimination and differential flux rates in maltoporin.
J.Mol.Biol., 272, 1997
|
|
2EZ8
 
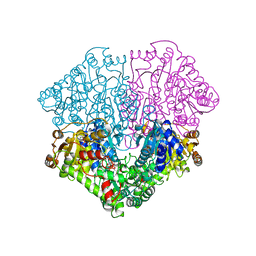 | | Pyruvate oxidase variant F479W in complex with reaction intermediate 2-lactyl-thiamin diphosphate | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Wille, G, Meyer, D, Steinmetz, A, Hinze, E, Golbik, R, Tittmann, K. | | Deposit date: | 2005-11-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.963 Å) | | Cite: | The catalytic cycle of a thiamin diphosphate enzyme examined by cryocrystallography.
Nat.Chem.Biol., 2, 2006
|
|
1LZJ
 
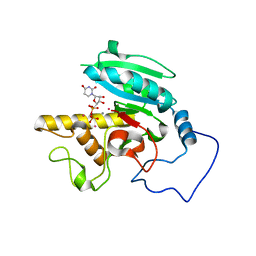 | | Glycosyltransferase B + UDP + H antigen acceptor | | Descriptor: | Glycosyltransferase B, MANGANESE (II) ION, MERCURY (II) ION, ... | | Authors: | Patenaude, S.I, Seto, N.O.L, Borisova, S.N, Szpacenko, A, Marcus, S.L, Palcic, M.M, Evans, S.V. | | Deposit date: | 2002-06-10 | | Release date: | 2002-08-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The structural basis for specificity in human
ABO(H) blood group biosynthesis.
Nat.Struct.Biol., 9, 2002
|
|
2EVU
 
 | | Crystal structure of aquaporin AqpM at 2.3A resolution | | Descriptor: | Aquaporin aqpM, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Lee, J.K, Kozono, D, Remis, J, Kitagawa, Y, Agre, P, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2005-10-31 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 A.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1MDW
 
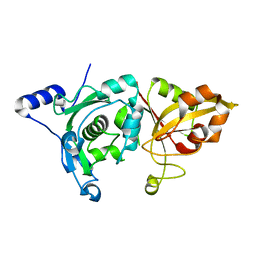 | | Crystal Structure of Calcium-Bound Protease Core of Calpain II Reveals the Basis for Intrinsic Inactivation | | Descriptor: | CALCIUM ION, Calpain II, catalytic subunit | | Authors: | Moldoveanu, T, Hosfield, C.M, Lim, D, Jia, Z, Davies, P.L. | | Deposit date: | 2002-08-07 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Calpain silencing by a reversible intrinsic mechanism.
Nat.Struct.Biol., 10, 2003
|
|
1S20
 
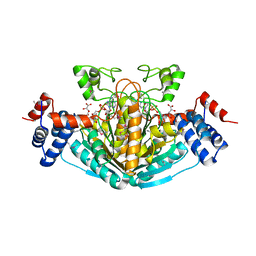 | | A novel NAD binding protein revealed by the crystal structure of E. Coli 2,3-diketogulonate reductase (YiaK) NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET ER82 | | Descriptor: | Hypothetical oxidoreductase yiaK, L(+)-TARTARIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Forouhar, F, Lee, I, Benach, J, Kulkarni, K, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-01-07 | | Release date: | 2004-01-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Novel NAD-binding Protein Revealed by the Crystal Structure of 2,3-Diketo-L-gulonate Reductase (YiaK).
J.Biol.Chem., 279, 2004
|
|
2F9B
 
 | | Discovery of Novel Heterocyclic Factor VIIa Inhibitors | | Descriptor: | Coagulation factor VII, Tissue factor, {5-(5-AMINO-1H-PYRROLO[3,2-B]PYRIDIN-2-YL)-6-HYDROXY-3'-NITRO-BIPHENYL-3-YL]-ACETIC ACID | | Authors: | Rai, R, Kolesnikov, A, Sprengeler, P.A, Torkelson, S, Ton, T, Katz, B.A, Yu, C, Hendrix, J, Shrader, W.D, Stephens, R, Cabuslay, R, Sanford, E, Young, W.B. | | Deposit date: | 2005-12-05 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Discovery of novel heterocyclic factor VIIa inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1MP6
 
 | |
1MPG
 
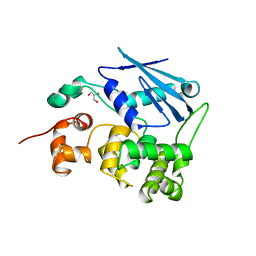 | | 3-METHYLADENINE DNA GLYCOSYLASE II FROM ESCHERICHIA COLI | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE II, GLYCEROL | | Authors: | Labahn, J, Schaerer, O.D, Long, A, Ezaz-Nikpay, K, Verdine, G.L, Ellenberger, T.E. | | Deposit date: | 1997-10-28 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the excision repair of alkylation-damaged DNA.
Cell(Cambridge,Mass.), 86, 1996
|
|
1MH9
 
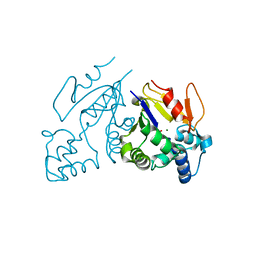 | | Crystal Structure Analysis of deoxyribonucleotidase | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, deoxyribonucleotidase | | Authors: | Rinaldo-Matthis, A, Rampazzo, C, Reichard, P, Bianchi, V, Nordlund, P. | | Deposit date: | 2002-08-19 | | Release date: | 2002-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a human mitochondrial deoxyribonucleotidase.
Nat.Struct.Biol., 9, 2002
|
|
1RY8
 
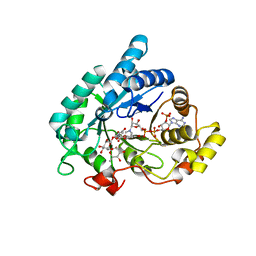 | | Prostaglandin F synthase complexed with NADPH and rutin | | Descriptor: | Aldo-keto reductase family 1 member C3, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, RUTIN | | Authors: | Komoto, J, Yamada, T, Watanabe, K, Takusagawa, F. | | Deposit date: | 2003-12-19 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of human prostaglandin F synthase (AKR1C3).
Biochemistry, 43, 2004
|
|
1MHS
 
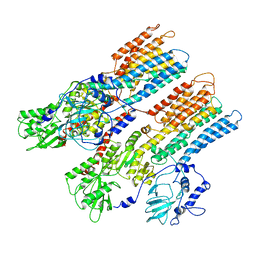 | | Model of Neurospora crassa proton ATPase | | Descriptor: | Plasma Membrane ATPase | | Authors: | Kuhlbrandt, W. | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | ELECTRON CRYSTALLOGRAPHY (8 Å) | | Cite: | Structure, mechanism and regulation of the Neurospora plasma membrane H+-ATPase
Science, 297, 2002
|
|
2FC0
 
 | | WRN exonuclease, Mn dGMP complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, Werner syndrome helicase | | Authors: | Perry, J.J, Tainer, J.A. | | Deposit date: | 2005-12-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2FCV
 
 | | SyrB2 with Fe(II), bromide, and alpha-ketoglutarate | | Descriptor: | ((2R,3S,4S,5S)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)-5-((2R,3S,4S,5S,6R)-3,4,5-TRIHYDROXY-6-METHOXY-TETRAHYDRO-2H-PYRAN-2-YLOXY)-TETRAHYDROFURAN-2-YL)METHYL NONANOATE, 2-OXOGLUTARIC ACID, BROMIDE ION, ... | | Authors: | Blasiak, L.C, Drennan, C.L. | | Deposit date: | 2005-12-12 | | Release date: | 2006-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the non-haem iron halogenase SyrB2 in syringomycin biosynthesis.
Nature, 440, 2006
|
|
1RXK
 
 | | crystal structure of streptavidin mutant (M3) a combination of M1+M2 | | Descriptor: | 5-(2-OXO-HEXAHYDRO-THIENO[3,4-D]IMIDAZOL-6-YL)-PENTANOIC ACID (4-NITRO-PHENYL)-AMIDE, Streptavidin | | Authors: | Eisenberg-Domovich, Y, Pazy, Y, Nir, O, Raboy, B, Bayer, E.A, Wilchek, M, Livnah, O. | | Deposit date: | 2003-12-18 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural elements responsible for conversion of streptavidin to a pseudoenzyme.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1S1T
 
 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with UC-781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, PHOSPHATE ION, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
2FF3
 
 | | Crystal structure of Gelsolin domain 1:N-wasp V2 motif hybrid in complex with actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Xue, B, Aguda, A.H, Robinson, R.C. | | Deposit date: | 2005-12-19 | | Release date: | 2006-03-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structural Basis of Actin Interaction with Multiple WH2/beta-Thymosin Motif-Containing Proteins
Structure, 14, 2006
|
|
1S06
 
 | | Crystal Structure of the R253K Mutant of 7,8-Diaminopelargonic Acid Synthase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, SODIUM ION | | Authors: | Sandmark, J, Eliot, A.C, Famm, K, Schneider, G, Kirsch, J.F. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from Escherichia coli are essential for catalysis.
Biochemistry, 43, 2004
|
|
1LXL
 
 | | NMR STRUCTURE OF BCL-XL, AN INHIBITOR OF PROGRAMMED CELL DEATH, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BCL-XL | | Authors: | Muchmore, S.W, Sattler, M, Liang, H, Meadows, R.P, Harlan, J.E, Yoon, H.S, Nettesheim, D, Chang, B.S, Thompson, C.B, Wong, S.L, Ng, S.C, Fesik, S.W. | | Deposit date: | 1996-04-04 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death.
Nature, 381, 1996
|
|
1S1R
 
 | | Crystal structures of prostaglandin D2 11-ketoreductase (AKR1C3) in complex with the non-steroidal anti-inflammatory drugs flufenamic acid and indomethacin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Aldo-keto reductase family 1 member C3, ... | | Authors: | Lovering, A.L, Ride, J.P, Bunce, C.M, Desmond, J.C, Cummings, S.M, White, S.A. | | Deposit date: | 2004-01-07 | | Release date: | 2004-03-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of prostaglandin D(2) 11-ketoreductase (AKR1C3) in complex with the nonsteroidal anti-inflammatory drugs flufenamic acid and indomethacin.
Cancer Res., 64, 2004
|
|
1S2A
 
 | | Crystal structures of prostaglandin D2 11-ketoreductase in complex with the non-steroidal anti-inflammatory drugs flufenamic acid and indomethacin | | Descriptor: | Aldo-keto reductase family 1 member C3, DIMETHYL SULFOXIDE, INDOMETHACIN, ... | | Authors: | Lovering, A.L, Ride, J.P, Bunce, C.M, Desmond, J.C, Cummings, S.M, White, S.A. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of prostaglandin D(2) 11-ketoreductase (AKR1C3) in complex with the nonsteroidal anti-inflammatory drugs flufenamic acid and indomethacin.
Cancer Res., 64, 2004
|
|
