4O65
 
 | | Crystal structure of the cupredoxin domain of amoB from Nitrosocaldus yellowstonii | | Descriptor: | COPPER (II) ION, Putative archaeal ammonia monooxygenase subunit B, SULFATE ION | | Authors: | Lawton, T.J, Ham, J, Sun, T, Rosenzweig, A.C. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-02 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Structural conservation of the B subunit in the ammonia monooxygenase/particulate methane monooxygenase superfamily.
Proteins, 82, 2014
|
|
7M4M
 
 | |
7M4N
 
 | |
7M4O
 
 | |
4F5J
 
 | |
4F5F
 
 | |
4F5M
 
 | |
4F5I
 
 | |
4F5H
 
 | |
8XXA
 
 | |
8XX9
 
 | | Rhodothermus marinus alpha-amylase RmGH13_47A CBM48-A-B-C domains | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Tonozuka, T. | | Deposit date: | 2024-01-18 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the recognition of alpha-1,6-branched alpha-glucan by GH13_47 alpha-amylase from Rhodothermus marinus
Proteins, 2024
|
|
8EB0
 
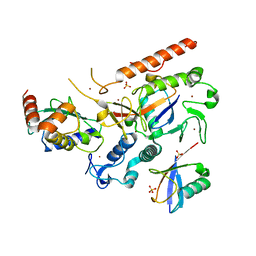 | | RNF216/E2-Ub/Ub transthiolation complex | | Descriptor: | E3 ubiquitin-protein ligase RNF216, SULFATE ION, Ubiquitin, ... | | Authors: | Cotton, T.R, Wang, X.S, Lechtenberg, B.C. | | Deposit date: | 2022-08-30 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | The unifying catalytic mechanism of the RING-between-RING E3 ubiquitin ligase family.
Nat Commun, 14, 2023
|
|
6XK9
 
 | | Cereblon in complex with DDB1, CC-90009, and GSPT1 | | Descriptor: | 2-(4-chlorophenyl)-N-({2-[(3S)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-5-yl}methyl)-2,2-difluoroacetamide, DNA damage-binding protein 1, Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, ... | | Authors: | Clayton, T.L, Tran, E.T, Zhu, J, Pagarigan, B.E, Matyskiela, M.E, Chamberlain, P.P. | | Deposit date: | 2020-06-25 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | CC-90009, a novel cereblon E3 ligase modulator, targets acute myeloid leukemia blasts and leukemia stem cells.
Blood, 137, 2021
|
|
4R0R
 
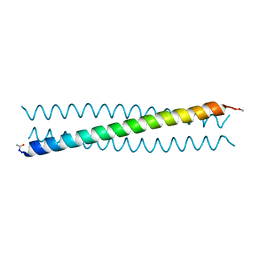 | | Ebolavirus GP Prehairpin Intermediate Mimic | | Descriptor: | eboIZN21 | | Authors: | Clinton, T.R, Weinstock, M.T, Jacobsen, M.T, Szabo-Fresnais, N, Pandya, M.J, Whitby, F.G, Herbert, A.S, Prugar, L.I, McKinnon, R, Hill, C.P, Welch, B.D, Dye, J.M, Eckert, D.M, Kay, M.S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-10-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design and characterization of ebolavirus GP prehairpin intermediate mimics as drug targets.
Protein Sci., 24, 2015
|
|
6NYR
 
 | |
6NYS
 
 | |
8FEI
 
 | |
8FEH
 
 | |
6M0D
 
 | | Beijerinckia indica beta-fructosyltransferase | | Descriptor: | Levansucrase, MAGNESIUM ION | | Authors: | Tonozuka, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 68 beta-fructosyltransferase from Beijerinckia indica subsp. indica in complex with fructose.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
6M0E
 
 | | Beijerinckia indica beta-fructosyltransferase complexed with fructose | | Descriptor: | Levansucrase, MAGNESIUM ION, beta-D-fructofuranose, ... | | Authors: | Tonozuka, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 68 beta-fructosyltransferase from Beijerinckia indica subsp. indica in complex with fructose.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
5NTU
 
 | | Crystal Structure of human Pro-myostatin Precursor at 2.6 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Growth/differentiation factor 8 | | Authors: | Cotton, T.R, Fischer, G, Hyvonen, M. | | Deposit date: | 2017-04-28 | | Release date: | 2018-01-17 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of the human myostatin precursor and determinants of growth factor latency.
EMBO J., 37, 2018
|
|
5NXS
 
 | |
4L6P
 
 | |
4L60
 
 | |
5MSE
 
 | | GFP nuclear transport receptor mimic 3B8 | | Descriptor: | Green fluorescent protein, IMIDAZOLE, SODIUM ION | | Authors: | Huyton, T, Gorlich, D. | | Deposit date: | 2017-01-04 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Surface Properties Determining Passage Rates of Proteins through Nuclear Pores.
Cell, 174, 2018
|
|
