8K9Z
 
 | | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-bound calmodulin | | Descriptor: | CALCIUM ION, Calmodulin-2, RDTND-RID CBD | | Authors: | Lee, Y, Choi, S, Hwang, J, Kim, M.H. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dissemination of pathogenic bacteria is reinforced by a MARTX toxin effector duet.
Nat Commun, 15, 2024
|
|
8KA1
 
 | | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-free calmodulin | | Descriptor: | Calmodulin-2, MAGNESIUM ION, RDTND-RID CBD | | Authors: | Lee, Y, Choi, S, Hwang, J, Kim, M.H. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Dissemination of pathogenic bacteria is reinforced by a MARTX toxin effector duet.
Nat Commun, 15, 2024
|
|
8KA0
 
 | | Crystal structure of Vibrio vulnificus RID-dependent transforming NADase domain (RDTND)/calmodulin-binding domain of Rho inactivation domain (RID-CBD) complexed with Ca2+-bound calmodulin and a nicotinamide adenine dinucleotide (NAD+) | | Descriptor: | CALCIUM ION, Calmodulin-2, GLYCEROL, ... | | Authors: | Lee, Y, Choi, S, Hwang, J, Kim, M.H. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Dissemination of pathogenic bacteria is reinforced by a MARTX toxin effector duet.
Nat Commun, 15, 2024
|
|
2BR9
 
 | | 14-3-3 Protein Epsilon (Human) Complexed to Peptide | | Descriptor: | 14-3-3 PROTEIN EPSILON, CONSENSUS PEPTIDE FOR 14-3-3 PROTEINS | | Authors: | Yang, X, Elkins, J.M, Soundararajan, M, Fedorov, O, Sundstrom, M, Edwards, A, Arrowsmith, C, Doyle, D.A. | | Deposit date: | 2005-05-03 | | Release date: | 2005-05-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Protein-Protein Interactions in the 14-3-3 Protein Family.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
5WSU
 
 | | Crystal structure of Myosin VIIa IQ5-SAH in complex with apo-CaM | | Descriptor: | Calmodulin, Unconventional myosin-VIIa | | Authors: | Li, J, Chen, Y, Deng, Y, Lu, Q, Zhang, M. | | Deposit date: | 2016-12-08 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Ca(2+)-Induced Rigidity Change of the Myosin VIIa IQ Motif-Single alpha Helix Lever Arm Extension
Structure, 25, 2017
|
|
4BW8
 
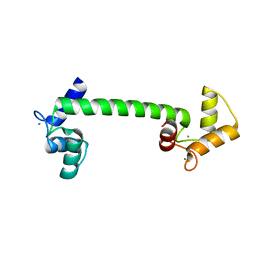 | | Calmodulin with small bend in central helix | | Descriptor: | CALCIUM ION, CALMODULIN | | Authors: | Kursula, P. | | Deposit date: | 2013-06-30 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Snapshots of Initial Steps in the Collapse of the Calmodulin Central Helix
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7WR3
 
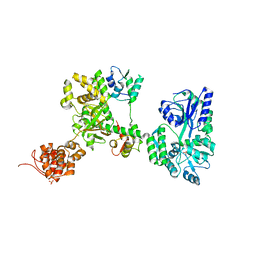 | | Crystal structure of MBP-fused OspC3 in complex with calmodulin | | Descriptor: | Calmodulin-1, MBP-fused OspC3, NICOTINAMIDE, ... | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6SZ5
 
 | |
7EEA
 
 | |
4UPU
 
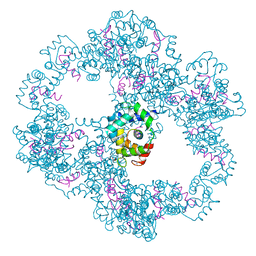 | | Crystal structure of IP3 3-K calmodulin binding region in complex with Calmodulin | | Descriptor: | CALCIUM ION, CALMODULIN, GLYCEROL, ... | | Authors: | Franco-Echevarria, E, Banos-Sanz, J.I, Monterroso, B, Round, A, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A New Calmodulin Binding Motif for Inositol 1,4,5-Trisphosphate 3-Kinase Regulation.
Biochem.J., 463, 2014
|
|
7E7F
 
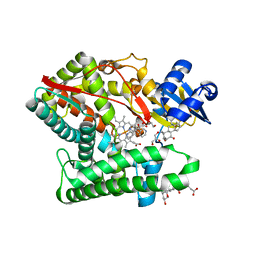 | | Human CYP11B1 mutant in complex with metyrapone | | Descriptor: | CHOLIC ACID, Cytochrome P450 11B1, mitochondrial, ... | | Authors: | Mukai, K, Sugimoto, H, Reiko, S, Matsuura, T, Hishiki, T, Kagawa, N. | | Deposit date: | 2021-02-26 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Spatially restricted substrate-binding site of cortisol-synthesizing CYP11B1 limits multiple hydroxylations and hinders aldosterone synthesis.
Curr Res Struct Biol, 3, 2021
|
|
4E34
 
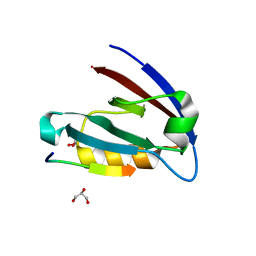 | | Crystal structure of CFTR Associated Ligand (CAL) PDZ domain bound to iCAL36 (ANSRWPTSII) peptide | | Descriptor: | GLYCEROL, Golgi-associated PDZ and coiled-coil motif-containing protein, decameric peptide, ... | | Authors: | Amacher, J.F, Beck, T, Madden, D.R. | | Deposit date: | 2012-03-09 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Stereochemical Determinants of C-terminal Specificity in PDZ Peptide-binding Domains: A NOVEL CONTRIBUTION OF THE CARBOXYLATE-BINDING LOOP.
J.Biol.Chem., 288, 2013
|
|
4E3B
 
 | |
7WR4
 
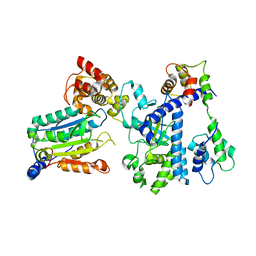 | | Crystal structure of OspC3-calmodulin-caspase-4 complex | | Descriptor: | Calmodulin-1, Caspase-4, OspC3 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5WSV
 
 | | Crystal structure of Myosin VIIa IQ5 in complex with Ca2+-CaM | | Descriptor: | CALCIUM ION, Calmodulin, SULFATE ION, ... | | Authors: | Li, J, Chen, Y, Deng, Y, Lu, Q, Zhang, M. | | Deposit date: | 2016-12-08 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Ca(2+)-Induced Rigidity Change of the Myosin VIIa IQ Motif-Single alpha Helix Lever Arm Extension
Structure, 25, 2017
|
|
3C36
 
 | |
1BAH
 
 | | A TWO DISULFIDE DERIVATIVE OF CHARYBDOTOXIN WITH DISULFIDE 13-33 REPLACED BY TWO ALPHA-AMINOBUTYRIC ACIDS, NMR, 30 STRUCTURES | | Descriptor: | CHARYBDOTOXIN | | Authors: | Song, J, Gilquin, B, Jamin, N, Guenneugues, M, Dauplais, M, Vita, C, Menez, A. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2020-01-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a two-disulfide derivative of charybdotoxin: structural evidence for conservation of scorpion toxin alpha/beta motif and its hydrophobic side chain packing.
Biochemistry, 36, 1997
|
|
4BW7
 
 | | Calmodulin in complex with strontium | | Descriptor: | CALMODULIN, STRONTIUM ION | | Authors: | Kursula, P. | | Deposit date: | 2013-06-30 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystallographic Snapshots of Initial Steps in the Collapse of the Calmodulin Central Helix
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7YV9
 
 | |
2W73
 
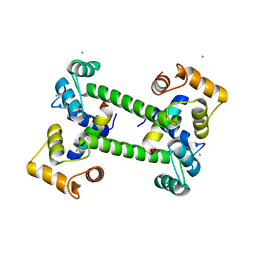 | |
4EUH
 
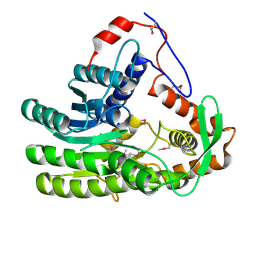 | | Crystal structure of Clostridium acetobutulicum trans-2-enoyl-CoA reductase apo form | | Descriptor: | Putative reductase CA_C0462, SODIUM ION | | Authors: | Hu, K, Zhao, M, Zhang, T, Yang, S, Ding, J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of trans-2-enoyl-CoA reductases from Clostridium acetobutylicum and Treponema denticola: insights into the substrate specificity and the catalytic mechanism
Biochem.J., 449, 2013
|
|
2WYS
 
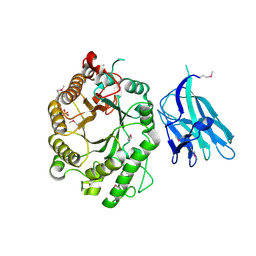 | | High resolution crystallographic structure of the Clostridium thermocellum N-terminal endo-1,4-beta-D-xylanase 10B (Xyn10B) CBM22-1- GH10 modules complexed with xylohexaose | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-XYLANASE Y, PHOSPHATE ION, ... | | Authors: | Najmudin, S, Pinheiro, B.A, Romao, M.J, Prates, J.A.M, Fontes, C.M.G.A. | | Deposit date: | 2009-11-20 | | Release date: | 2010-08-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Putting an N-Terminal End to the Clostridium Thermocellum Xylanase Xyn10B Story: Crystal Structure of the Cbm22-1-Gh10 Modules Complexed with Xylohexaose.
J.Struct.Biol., 172, 2010
|
|
2X0G
 
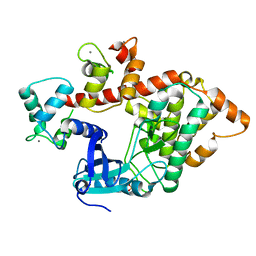 | | X-RAY STRUCTURE OF A DAP-KINASE CALMODULIN COMPLEX | | Descriptor: | CALCIUM ION, CALMODULIN, DEATH-ASSOCIATED PROTEIN KINASE 1, ... | | Authors: | Kuper, J, De Diego, I, Lehmann, F, Wilmanns, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Basis of the Death-Associated Protein Kinase-Calcium/Calmodulin Regulator Complex.
Sci.Signal, 3, 2010
|
|
2Y4V
 
 | |
4AV3
 
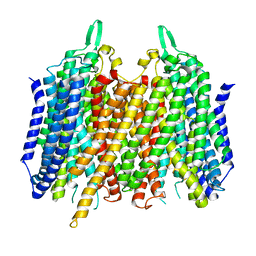 | | Crystal structure of Thermotoga Maritima sodium pumping membrane integral pyrophosphatase with metal ions in active site | | Descriptor: | CALCIUM ION, K(+)-STIMULATED PYROPHOSPHATE-ENERGIZED SODIUM PUMP, MAGNESIUM ION | | Authors: | Kajander, T, Kogan, K, Kellosalo, J, Pokharel, K, Goldman, A. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure and Catalytic Cycle of a Sodium-Pumping Pyrophosphatase.
Science, 337, 2012
|
|
