4MZG
 
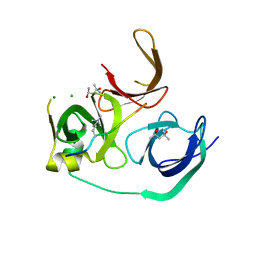 | | Crystal structure of human Spindlin1 bound to histone H3K4me3 peptide | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Su, X, Ding, X, Li, H. | | Deposit date: | 2013-09-30 | | Release date: | 2014-03-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Molecular basis underlying histone H3 lysine-arginine methylation pattern readout by Spin/Ssty repeats of Spindlin1
Genes Dev., 28, 2014
|
|
2V83
 
 | |
8FJN
 
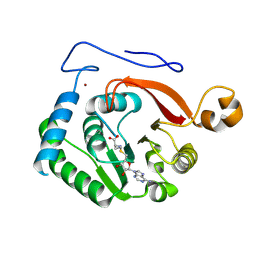 | | Crystal Structure of the Trypanosoma brucei DOT1A histone H3K76 methyltransferase in complex with AdoHcy - C2221 space group | | Descriptor: | CALCIUM ION, CHLORIDE ION, Histone-lysine N-methyltransferase, ... | | Authors: | Frisbie, V.S, Hashimoto, H, Debler, E.W. | | Deposit date: | 2022-12-20 | | Release date: | 2024-02-07 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two DOT1 enzymes cooperatively mediate efficient ubiquitin-independent histone H3 lysine 76 tri-methylation in kinetoplastids.
Nat Commun, 15, 2024
|
|
6JM9
 
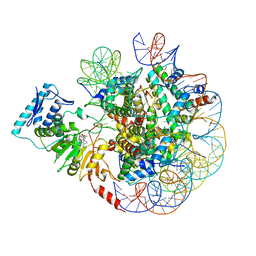 | | cryo-EM structure of DOT1L bound to unmodified nucleosome | | Descriptor: | DNA strand I, DNA strand J, Histone H2A, ... | | Authors: | Jang, S, Song, J.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structural basis of recognition and destabilization of the histone H2B ubiquitinated nucleosome by the DOT1L histone H3 Lys79 methyltransferase.
Genes Dev., 33, 2019
|
|
5VNB
 
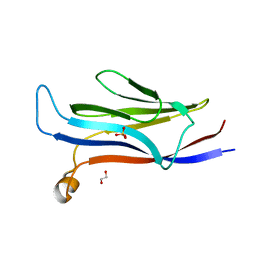 | | YEATS in complex with histone H3 | | Descriptor: | 1,2-ETHANEDIOL, H3K23acK27ac peptide, SULFATE ION, ... | | Authors: | Cho, H.J, Cierpicki, T. | | Deposit date: | 2017-04-29 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | GAS41 Recognizes Diacetylated Histone H3 through a Bivalent Binding Mode.
ACS Chem. Biol., 13, 2018
|
|
6JMA
 
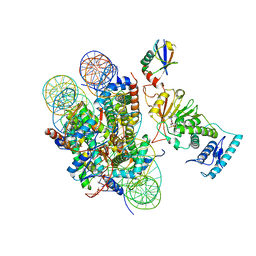 | | cryo-EM structure of DOT1L bound to H2B ubiquitinated nucleosome | | Descriptor: | DNA I&J, Histone H2A, Histone H2B 1.1, ... | | Authors: | Jang, S, Song, J.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structural basis of recognition and destabilization of the histone H2B ubiquitinated nucleosome by the DOT1L histone H3 Lys79 methyltransferase.
Genes Dev., 33, 2019
|
|
8COK
 
 | |
4GNG
 
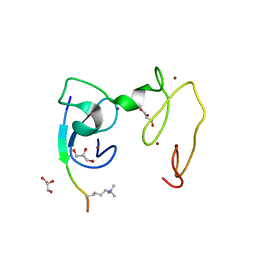 | | Crystal Structure of NSD3 tandem PHD5-C5HCH domains complexed with H3K9me3 peptide | | Descriptor: | GLYCEROL, Histone H3.3, Histone-lysine N-methyltransferase NSD3, ... | | Authors: | Li, F, He, C, Wu, J, Shi, Y. | | Deposit date: | 2012-08-17 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The methyltransferase NSD3 has chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone H3 recognition.
J.Biol.Chem., 288, 2013
|
|
3K24
 
 | | Crystal structure of mature apo-Cathepsin L C25A mutant in complex with Gln-Leu-Ala peptide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-6)-beta-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-2)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cathepsin L1, ... | | Authors: | Adams-Cioaba, M.A, Krupa, J.C, Mort, J.S, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-29 | | Release date: | 2010-03-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the recognition and cleavage of histone H3 by cathepsin L.
Nat Commun, 2, 2011
|
|
1ML9
 
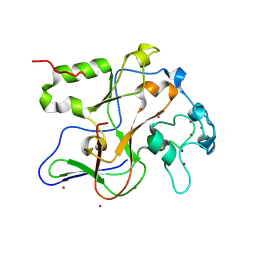 | | Structure of the Neurospora SET domain protein DIM-5, a histone lysine methyltransferase | | Descriptor: | Histone H3 methyltransferase DIM-5, UNKNOWN LIGAND, ZINC ION | | Authors: | Zhang, X, Tamaru, H, Khan, S.I, Horton, J.R, Keefe, L.J, Selker, E.U, Cheng, X. | | Deposit date: | 2002-08-30 | | Release date: | 2002-10-23 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the Neurospora SET domain protein DIM-5,
a histone H3 lysine methyltransferase
Cell(Cambridge,Mass.), 111, 2002
|
|
2LGG
 
 | | Structure of PHD domain of UHRF1 in complex with H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION, histone H3 peptide | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-26 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
5JIY
 
 | | Structure of G9a SET-domain with Histone H3K9norLeucine mutant peptide and bound S-adenosylmethionine | | Descriptor: | Histone H3.1 mutant peptide with H3K9nor-leucine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Jayaram, H, Bellon, S.F, Poy, F. | | Deposit date: | 2016-04-22 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JHN
 
 | | Structure of G9a SET-domain with Histone H3K9Ala mutant peptide and bound S-adenosylmethionine | | Descriptor: | Histone H3.1 peptide with K9A mutation, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Jayaram, H, Bellon, S.F, Poy, F. | | Deposit date: | 2016-04-21 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JIN
 
 | | Structure of G9a SET-domain with Histone H3K9M mutant peptide and bound S-adenosylmethionine | | Descriptor: | Histone H3.1 peptide with K9M mutation, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYLMETHIONINE, ... | | Authors: | Jayaram, H, Bellon, S.F, Poy, F. | | Deposit date: | 2016-04-22 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2RVN
 
 | |
3G7L
 
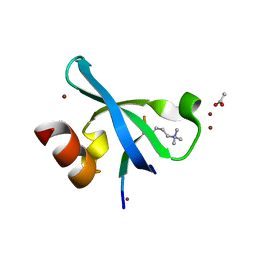 | | Chromodomain of Chp1 in complex with Histone H3K9me3 peptide | | Descriptor: | ACETIC ACID, Chromo domain-containing protein 1, Histone H3.1/H3.2, ... | | Authors: | Schalch, T, Joshua-Tor, L. | | Deposit date: | 2009-02-10 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin
Mol.Cell, 34, 2009
|
|
5CMA
 
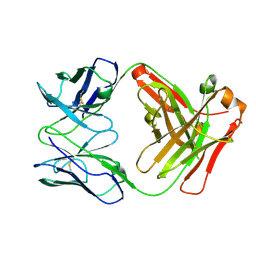 | | Anti-B7-H3 monoclonal antibody ch8H9 Fab fragment | | Descriptor: | Antibody ch8H9 Fab heavy chain, Antibody ch8H9 Fab light chain | | Authors: | Ahmed, M, Goldgur, Y, Cheng, M, Cheung, N.K. | | Deposit date: | 2015-07-16 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Humanized Affinity-matured Monoclonal Antibody 8H9 Has Potent Antitumor Activity and Binds to FG Loop of Tumor Antigen B7-H3.
J.Biol.Chem., 290, 2015
|
|
2H68
 
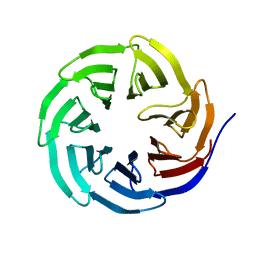 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | Descriptor: | WD-repeat protein 5 | | Authors: | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-30 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2DZE
 
 | |
9BDF
 
 | |
9BDG
 
 | |
4I0K
 
 | | Crystal structure of murine B7-H3 extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CD276 antigen, SULFATE ION, ... | | Authors: | Vigdorovich, V, Ramagopal, U, Bonanno, J.B, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-11-16 | | Release date: | 2012-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structure and T cell inhibition properties of B7 family member, B7-H3.
Structure, 21, 2013
|
|
3RJ2
 
 | |
2KVM
 
 | |
2G9A
 
 | |
