8OOR
 
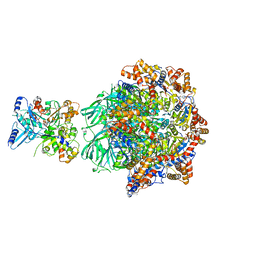 | | CryoEM Structure INO80core Hexasome complex Rvb core refinement state2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 5, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8PKI
 
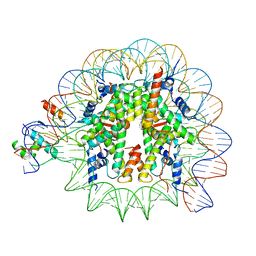 | | Cryo-EM structure of NR5A2-nucleosome complex SHL+5.5 | | Descriptor: | DNA, Histone H2A, Histone H2B type 1-C/E/G, ... | | Authors: | Kobayashi, W, Sappler, A, Bollschweiler, D, Kummecke, M, Basquin, J, Arslantas, E, Ruangroengkulrith, S, Hornberger, R, Duderstadt, K, Tachibana, K. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8PKJ
 
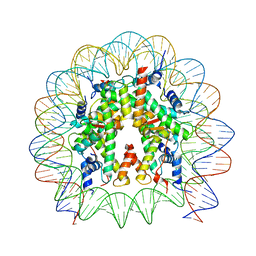 | | Cryo-EM structure of the nucleosome containing Nr5a2 motif at SHL+5.5 | | Descriptor: | DNA, Histone H2A, Histone H2B, ... | | Authors: | Kobayashi, W, Sappler, A, Bollschweiler, D, Kummecke, M, Basquin, J, Arslantas, E, Ruangroengkulrith, S, Hornberger, R, Duderstadt, K, Tachibana, K. | | Deposit date: | 2023-06-26 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8PQY
 
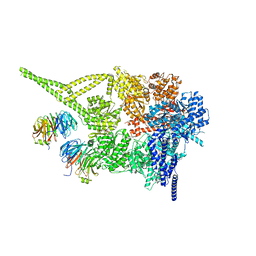 | | Cytoplasmic dynein-1 motor domain bound to LIS1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1, ... | | Authors: | Singh, K, Lau, C.K, Manigrasso, G, Gassmann, R, Carter, A.P. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular mechanism of dynein-dynactin complex assembly by LIS1.
Science, 383, 2024
|
|
8PR3
 
 | | Cytoplasmic dynein-1 heavy chain bound to JIP3-RH1 | | Descriptor: | C-Jun-amino-terminal kinase-interacting protein 3, Cytoplasmic dynein 1 heavy chain 1, Cytoplasmic dynein 1 intermediate chain 2, ... | | Authors: | Singh, K, Lau, C.K, Manigrasso, G, Gassmann, R, Carter, A.P. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular mechanism of dynein-dynactin complex assembly by LIS1.
Science, 383, 2024
|
|
8PR2
 
 | | Cytoplasmic dynein-1 heavy chain bound to JIP3-LZI | | Descriptor: | C-Jun-amino-terminal kinase-interacting protein 3, Cytoplasmic dynein 1 heavy chain 1, Cytoplasmic dynein 1 intermediate chain 2, ... | | Authors: | Singh, K, Lau, C.K, Manigrasso, G, Gassmann, R, Carter, A.P. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular mechanism of dynein-dynactin complex assembly by LIS1.
Science, 383, 2024
|
|
8PQW
 
 | | Cytoplasmic dynein-1 motor domain bound to dynactin-p150glued and LIS1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1, ... | | Authors: | Singh, K, Lau, C.K, Manigrasso, G, Gassmann, R, Carter, A.P. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular mechanism of dynein-dynactin complex assembly by LIS1.
Science, 383, 2024
|
|
8PR0
 
 | | Cytoplasmic dynein-A heavy chain bound to dynactin-p150glued and IC-LC tower | | Descriptor: | Cytoplasmic dynein 1 heavy chain 1, Cytoplasmic dynein 1 intermediate chain 2, Cytoplasmic dynein 1 light intermediate chain 2, ... | | Authors: | Singh, K, Lau, C.K, Manigrasso, G, Gassmann, R, Carter, A.P. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Molecular mechanism of dynein-dynactin complex assembly by LIS1.
Science, 383, 2024
|
|
8PR1
 
 | | Cytoplasmic dynein-B heavy chain bound to IC-LC tower | | Descriptor: | Cytoplasmic dynein 1 heavy chain 1, Cytoplasmic dynein 1 intermediate chain 2, Cytoplasmic dynein 1 light intermediate chain 2, ... | | Authors: | Singh, K, Lau, C.K, Manigrasso, G, Gassmann, R, Carter, A.P. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Molecular mechanism of dynein-dynactin complex assembly by LIS1.
Science, 383, 2024
|
|
8PQZ
 
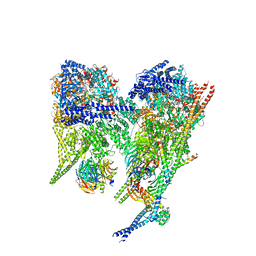 | | Cytoplasmic dynein-1 A1/A2 motor domains bound to LIS1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1, ... | | Authors: | Singh, K, Lau, C.K, Manigrasso, G, Gassmann, R, Carter, A.P. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Molecular mechanism of dynein-dynactin complex assembly by LIS1.
Science, 383, 2024
|
|
8PID
 
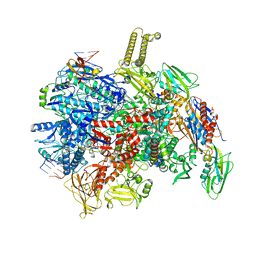 | | backtracked E. coli transcription complex paused at ops site and bound to RfaH | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuber, P.K, Said, N, Hilal, T, Loll, B, Wahl, M.C, Knauer, S.H. | | Deposit date: | 2023-06-21 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Concerted transformation of a hyper-paused transcription complex and its reinforcing protein.
Nat Commun, 15, 2024
|
|
8PFJ
 
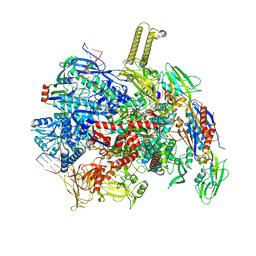 | | fully recruited RfaH bound to E. coli transcription complex paused at ops site (not fully complementary scaffold; alternative state of RfaH) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuber, P.K, Said, N, Hilal, T, Loll, B, Wahl, M.C, Knauer, S.H. | | Deposit date: | 2023-06-16 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Concerted transformation of a hyper-paused transcription complex and its reinforcing protein.
Nat Commun, 15, 2024
|
|
8PIL
 
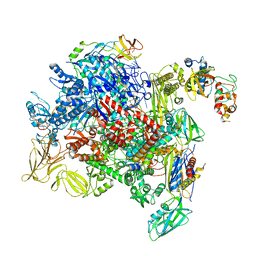 | | E. coli transcription complex paused at ops site and bound to RfaH and NusA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuber, P.K, Said, N, Hilal, T, Loll, B, Wahl, M.C, Knauer, S.H. | | Deposit date: | 2023-06-22 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Concerted transformation of a hyper-paused transcription complex and its reinforcing protein.
Nat Commun, 15, 2024
|
|
8OZ5
 
 | |
8P02
 
 | |
8OQX
 
 | | Crystal structure of Tannerella forsythia MurNAc kinase MurK with a phosphate analogue | | Descriptor: | 1,2-ETHANEDIOL, ATPase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gogler, K, Fink, P, Stasiak, A.C, Stehle, T, Zocher, G. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | N-acetylmuramic acid recognition by MurK kinase from the MurNAc auxotrophic oral pathogen Tannerella forsythia.
J.Biol.Chem., 299, 2023
|
|
8OIC
 
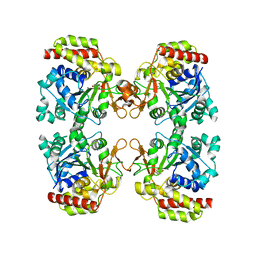 | | Trichomonas vaginalis riboside hydrolase (His-tagged) | | Descriptor: | BICINE, CALCIUM ION, Inosine-uridine preferring nucleoside hydrolase family protein | | Authors: | Patrone, M, Stockman, B.J, Degano, M. | | Deposit date: | 2023-03-22 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A riboside hydrolase that salvages both nucleobases and nicotinamide in the auxotrophic parasite Trichomonas vaginalis.
J.Biol.Chem., 299, 2023
|
|
8OOC
 
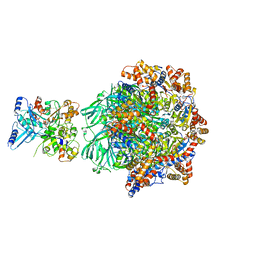 | | CryoEM Structure INO80core Hexasome complex Rvb core refinement state1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chromatin-remodeling ATPase Ino80, ... | | Authors: | Zhang, M, Jungblut, A, Hoffmann, T, Eustermann, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Hexasome-INO80 complex reveals structural basis of noncanonical nucleosome remodeling.
Science, 381, 2023
|
|
8OPN
 
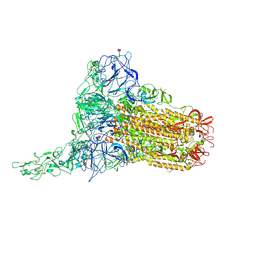 | | Human Coronavirus HKU1 spike glycoprotein in complex with an alpha2,8-linked 9-O-acetylated disialoside (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-8)-N-acetyl-alpha-neuraminic acid, ... | | Authors: | Pronker, M.F, Creutznacher, R, Hurdiss, D.L. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Sialoglycan binding triggers spike opening in a human coronavirus.
Nature, 624, 2023
|
|
8OPM
 
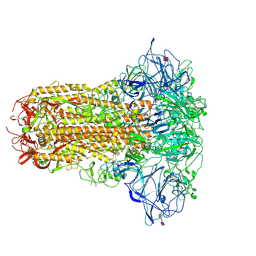 | | Human Coronavirus HKU1 spike glycoprotein in complex with an alpha2,8-linked 9-O-acetylated disialoside (closed state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-8)-N-acetyl-alpha-neuraminic acid, ... | | Authors: | Pronker, M.F, Creutznacher, R, Hurdiss, D.L. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Sialoglycan binding triggers spike opening in a human coronavirus.
Nature, 624, 2023
|
|
8OPO
 
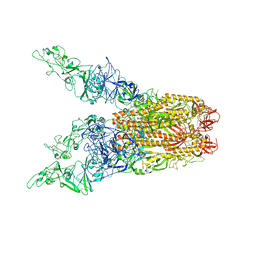 | | Human Coronavirus HKU1 spike glycoprotein in complex with an alpha2,8-linked 9-O-acetylated disialoside (3-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-8)-N-acetyl-alpha-neuraminic acid, ... | | Authors: | Pronker, M.F, Creutznacher, R, Hurdiss, D.L. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Sialoglycan binding triggers spike opening in a human coronavirus.
Nature, 624, 2023
|
|
8OW0
 
 | | Cryo-EM structure of CBF1-CCAN bound topologically to a centromeric CENP-A nucleosome | | Descriptor: | C0N3 DNA, Centromere-binding protein 1, Histone H2A.1, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
8PEW
 
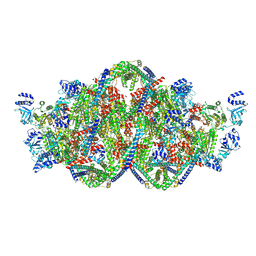 | | Rho-ATPgS-Psu complex II | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Polarity suppression protein, ... | | Authors: | Gjorgjevikj, D, Wahl, M.C, Hilal, T, Loll, B. | | Deposit date: | 2023-06-15 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Rho-ATPgS-Psu complex II
To Be Published
|
|
8PEL
 
 | |
8PEQ
 
 | | Complex of diubiquitin-derived artificial binding protein (Affilin) variant Af2 with its target oncofetal fibronectin (fragment 7B8) | | Descriptor: | Affilin variant Af2, Fibronectin, SULFATE ION | | Authors: | Parthier, C, Katzschmann, A, Fiedler, E, Haupts, U, Reimann, A. | | Deposit date: | 2023-06-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Ubiquitin-derived artificial binding proteins targeting oncofetal fibronectin reveal scaffold plasticity by beta-strand slippage.
Commun Biol, 7, 2024
|
|
