5NGS
 
 | | Crystal structure of human MTH1 in complex with inhibitor 6-[(2-phenylethyl)sulfanyl]-7H-purin-2-amine | | Descriptor: | 6-(2-phenylethylsulfanyl)-7~{H}-purin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, ... | | Authors: | Gustafsson, R, Rudling, A, Almlof, I, Homan, E, Scobie, M, Warpman Berglund, U, Helleday, T, Carlsson, J, Stenmark, P. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-Based Discovery and Optimization of Enzyme Inhibitors by Docking of Commercial Chemical Space.
J. Med. Chem., 60, 2017
|
|
7JJL
 
 | | Crystal structure of Importin Alpha 3 in complex with human LSD1 NLS | | Descriptor: | Importin subunit alpha-3, Lysine-specific histone demethylase 1A | | Authors: | Tu, W.J, McCuaig, R, Tan, H.Y.A, Hardy, K, Seddiki, N, Ali, S, Dahlstrom, J.E, Bean, E.G, Dunn, J, Forwood, J.K, Tsimbalyuk, S, Smith, K, Yip, D, Malik, L, Prasana, T, Milburn, P, Rao, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Targeting Nuclear LSD1 to Reprogram Cancer Cells and Reinvigorate Exhausted T Cells via a Novel LSD1-EOMES Switch.
Front Immunol, 11, 2020
|
|
6BXG
 
 | | 1.45 Angstrom Resolution Crystal Structure of PDZ domain of Carboxy-Terminal Protease from Vibrio cholerae in Complex with Peptide. | | Descriptor: | CHLORIDE ION, IODIDE ION, LEU-ILE-ALA, ... | | Authors: | Minasov, G, Shuvalova, L, Filippova, E.V, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-12-18 | | Release date: | 2018-01-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 Angstrom Resolution Crystal Structure of PDZ domain of Carboxy-Terminal Protease from Vibrio cholerae in Complex with Peptide.
To Be Published
|
|
4IXQ
 
 | | RT fs X-ray diffraction of Photosystem II, dark state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Alonso-Mori, R, Tran, R, Hattne, J, Gildea, R.J, Echols, N, Gloeckner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Koroidov, S, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, Schafer, D.W, Messerschmidt, M, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Grosse-Kunstleve, R.W, Zwart, P.H, White, W.E, Glatzel, P, Adams, P.D, Bogan, M.J, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Sauter, N.K, Yachandra, V.K, Bergmann, U, Yano, J. | | Deposit date: | 2013-01-27 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.7 Å) | | Cite: | Simultaneous femtosecond X-ray spectroscopy and diffraction of photosystem II at room temperature.
Science, 340, 2013
|
|
7KGE
 
 | |
7KGI
 
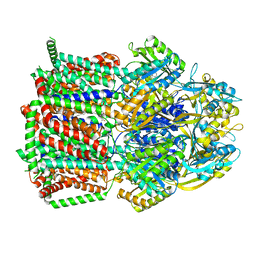 | |
7L30
 
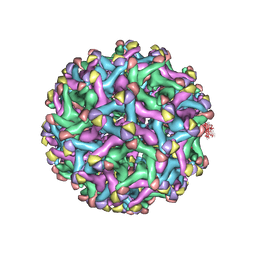 | | Binjari virus (BinJV) | | Descriptor: | Envelope protein E, prM protein | | Authors: | Hardy, J.M, Venugopal, H.V, Newton, N.D, Watterson, D, Coulibaly, F.J. | | Deposit date: | 2020-12-17 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The structure of an infectious immature flavivirus redefines viral architecture and maturation.
Sci Adv, 7, 2021
|
|
7KGH
 
 | |
5V91
 
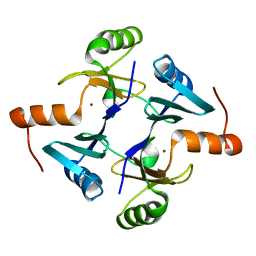 | | Crystal structure of fosfomycin resistance protein from Klebsiella pneumoniae | | Descriptor: | Fosfomycin resistance protein, ZINC ION | | Authors: | Klontz, E, Guenther, S, Silverstein, Z, Sundberg, E. | | Deposit date: | 2017-03-22 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and Dynamics of FosA-Mediated Fosfomycin Resistance in Klebsiella pneumoniae and Escherichia coli.
Antimicrob. Agents Chemother., 61, 2017
|
|
2X5V
 
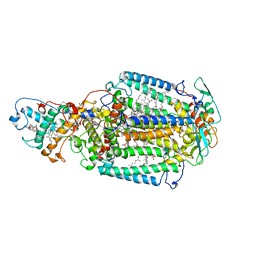 | | 80 microsecond laue diffraction snapshot from crystals of a photosynthetic reaction centre 3 millisecond following photoactivation. | | Descriptor: | BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, FE (II) ION, ... | | Authors: | Wohri, A.B, Katona, G, Johansson, L.C, Fritz, E, Malmerberg, E, Andersson, M, Vincent, J, Eklund, M, Cammarata, M, Wulff, M, Davidsson, J, Groenhof, G, Neutze, R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Light-Induced Structural Changes in a Photosynthetic Reaction Center Caught by Laue Diffraction.
Science, 328, 2010
|
|
7KJV
 
 | | Structure of HIV-1 reverse transcriptase initiation complex core | | Descriptor: | HIV-1 viral RNA fragment, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ha, B, Larsen, K.P, Zhang, J, Fu, Z, Montabana, E, Jackson, L.N, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-17 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | High-resolution view of HIV-1 reverse transcriptase initiation complexes and inhibition by NNRTI drugs.
Nat Commun, 12, 2021
|
|
5CPV
 
 | |
3KJV
 
 | | HIV-1 reverse transcriptase in complex with DNA | | Descriptor: | 5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*(DOC))-3', 5'-D(*AP*TP*GP*GP*TP*GP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2009-11-03 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Visualizing the molecular interactions of a nucleotide analog, GS-9148, with HIV-1 reverse transcriptase-DNA complex.
J.Mol.Biol., 397, 2010
|
|
2PIN
 
 | | Thyroid receptor beta in complex with inhibitor | | Descriptor: | 1-(4-HEXYLPHENYL)PROP-2-EN-1-ONE, SULFATE ION, Thyroid hormone receptor beta-1, ... | | Authors: | Estebanez-Perpina, E, Jouravel, N, Baxter, J.D, Guy, L.R, Webb, P, Fletterick, R.J. | | Deposit date: | 2007-04-13 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into the mode of action of a direct inhibitor of coregulator binding to the thyroid hormone receptor.
Mol.Endocrinol., 21, 2007
|
|
7KJW
 
 | | Structure of HIV-1 reverse transcriptase initiation complex core with efavirenz | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 viral RNA fragment, MAGNESIUM ION, ... | | Authors: | Ha, B, Larsen, K.P, Zhang, J, Fu, Z, Montabana, E, Jackson, L.N, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-17 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-resolution view of HIV-1 reverse transcriptase initiation complexes and inhibition by NNRTI drugs.
Nat Commun, 12, 2021
|
|
2WKF
 
 | | Crystal Structure of Macrophage Migration Inhibitory Factor from Plasmodium falciparum | | Descriptor: | GLYCEROL, MACROPHAGE MIGRATION INHIBITORY FACTOR | | Authors: | Dobson, S.E, Augustijn, K.D, Brannigan, J.A, Dodson, E.J, Waters, A.P, Wilkinson, A.J. | | Deposit date: | 2009-06-11 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The crystal structures of macrophage migration inhibitory factor from Plasmodium falciparum and Plasmodium berghei.
Protein Sci., 18, 2009
|
|
7D76
 
 | | Cryo-EM structure of the beclomethasone-bound adhesion receptor GPR97-Go complex | | Descriptor: | (8~{S},9~{R},10~{S},11~{S},13~{S},14~{S},16~{S},17~{R})-9-chloranyl-10,13,16-trimethyl-11,17-bis(oxidanyl)-17-(2-oxidanylethanoyl)-6,7,8,11,12,14,15,16-octahydrocyclopenta[a]phenanthren-3-one, Adhesion G protein-coupled receptor G3; GPR97, CHOLESTEROL, ... | | Authors: | Ping, Y, Mao, C, Xiao, P, Zhao, R, Jiang, Y, Yang, Z, An, W, Shen, D, Yang, F, Zhang, H, Qu, C, Shen, Q, Tian, C, Li, Z, Li, S, Wang, G, Tao, X, Wen, X, Zhong, Y, Yang, J, Yi, F, Yu, X, Xu, E, Zhang, Y, Sun, J. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the glucocorticoid-bound adhesion receptor GPR97-G o complex.
Nature, 589, 2021
|
|
7KGD
 
 | |
5H22
 
 | | Hsp90 alpha N-terminal domain in complex with an inhibitor | | Descriptor: | 4-chloranyl-7-[(4-methoxy-3,5-dimethyl-pyridin-2-yl)methyl]-5-(phenylmethyl)pyrrolo[2,3-d]pyrimidin-2-amine, Hsp90aa1 protein | | Authors: | Kim, E.E, Shin, S.C, Keum, G.C. | | Deposit date: | 2016-10-13 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Synthesis and in vitro antiproliferative activity of C5-benzyl substituted 2-amino-pyrrolo[2,3-d]pyrimidines as potent Hsp90 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7KGG
 
 | |
1ZLY
 
 | | The structure of human glycinamide ribonucleotide transformylase in complex with alpha,beta-N-(hydroxyacetyl)-D-ribofuranosylamine and 10-formyl-5,8,dideazafolate | | Descriptor: | 4-[(4-{[(2-AMINO-4-OXO-3,4-DIHYDROQUINAZOLIN-6-YL)METHYL]AMINO}BENZOYL)AMINO]BUTANOIC ACID, 5-O-phosphono-beta-D-ribofuranosylamine, Phosphoribosylglycinamide formyltransferase | | Authors: | Dahms, T.E.S, Sainz, G, Giroux, E.L, Caperelli, C.A, Smith, J.L. | | Deposit date: | 2005-05-09 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The apo and ternary complex structures of a chemotherapeutic target: human glycinamide ribonucleotide transformylase.
Biochemistry, 44, 2005
|
|
1EEI
 
 | |
2WMK
 
 | | Crystal structure of the catalytic module of a family 98 glycoside hydrolase from Streptococcus pneumoniae SP3-BS71 (Sp3GH98) in complex with the A-LewisY pentasaccharide blood group antigen. | | Descriptor: | FUCOLECTIN-RELATED PROTEIN, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Higgins, M.A, Whitworth, G.E, El Warry, N, Randriantsoa, M, Samain, E, Burke, R.D, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential Recognition and Hydrolysis of Host Carbohydrate-Antigens by Streptococcus Pneumoniae Family 98 Glycoside Hydrolases.
J.Biol.Chem., 284, 2009
|
|
5CXA
 
 | | Crystal structure of the catalytic domain of Human MMP12 in complex with a carboxylate inhibitor related to RXP470 | | Descriptor: | (R)-4-(((S)-1-(((S)-1-amino-4-carboxy-1-oxobutan-2-yl)amino)-4-carboxy-1-oxobutan-2-yl)amino)-3-((3-(3'-chloro-[1,1'-biphenyl]-4-yl)isoxazol-5-yl)methyl)-4-oxobutanoate, CALCIUM ION, Macrophage metalloelastase, ... | | Authors: | Rouanet-Mehouas, C, Devel, L, Dive, V, Stura, E.A. | | Deposit date: | 2015-07-28 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Zinc-Metalloproteinase Inhibitors: Evaluation of the Complex Role Played by the Zinc-Binding Group on Potency and Selectivity.
J. Med. Chem., 60, 2017
|
|
2AV0
 
 | | F97L with CO bound | | Descriptor: | CARBON MONOXIDE, Globin I, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Bonham, M.A, Gibson, Q.H, Nichols, J.C, Royer Jr, W.E. | | Deposit date: | 2005-08-29 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Residue F4 plays a key role in modulating oxygen affinity and cooperativity in Scapharca dimeric hemoglobin
Biochemistry, 44, 2005
|
|
