7NM1
 
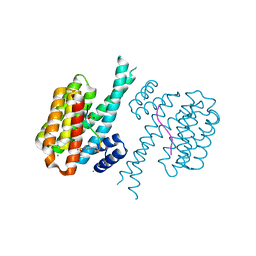 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-126 | | Descriptor: | 14-3-3 protein sigma, 4-(4-pyrrolidin-1-ylpiperidin-1-yl)sulfonylbenzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7NJB
 
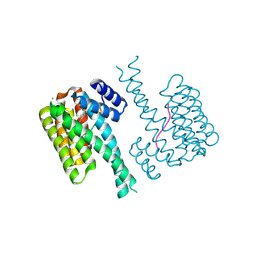 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-132 | | Descriptor: | 14-3-3 protein sigma, 4-piperidin-1-ylsulfonylbenzaldehyde, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7NK3
 
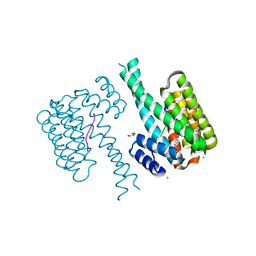 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-128 | | Descriptor: | 14-3-3 protein sigma, 2-(4-methylphenyl)sulfonyl-3,4-dihydro-1~{H}-isoquinoline, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-02-17 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7NLA
 
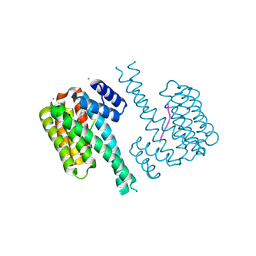 | |
4NMO
 
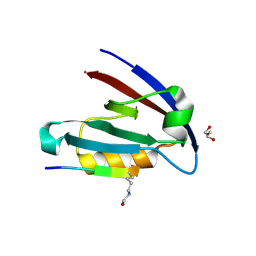 | |
4NLC
 
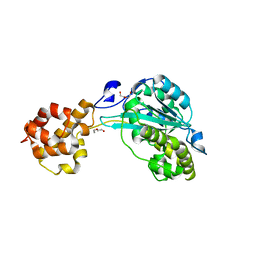 | |
7NEH
 
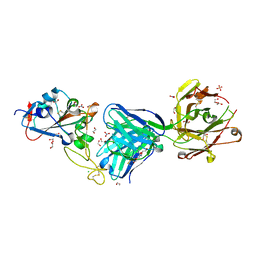 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-269 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-02-04 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera.
Cell, 184, 2021
|
|
4ESE
 
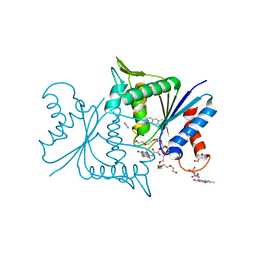 | | The crystal structure of azoreductase from Yersinia pestis CO92 in complex with FMN. | | Descriptor: | DODECAETHYLENE GLYCOL, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase, ... | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The crystal structure of acyl carrier protein phosphodiesterase from Yersinia pestis CO92 in complex with FMN.
To be Published
|
|
4O0J
 
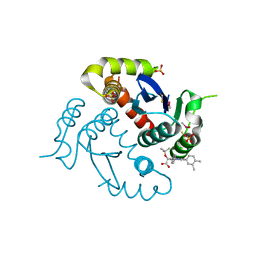 | | HIV-1 Integrase Catalytic Core Domain Complexed with Allosteric Inhibitor (2S)-tert-butoxy[4-(4-chlorophenyl)-6-(3,4-dimethylphenyl)-2,5-dimethylpyridin-3-yl]ethanoic acid | | Descriptor: | (2S)-tert-butoxy[4-(4-chlorophenyl)-6-(3,4-dimethylphenyl)-2,5-dimethylpyridin-3-yl]ethanoic acid, Integrase, SULFATE ION | | Authors: | Feng, L, Kvaratskhelia, M. | | Deposit date: | 2013-12-13 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A New Class of Multimerization Selective Inhibitors of HIV-1 Integrase.
Plos Pathog., 10, 2014
|
|
6SGC
 
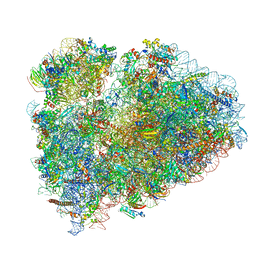 | | Rabbit 80S ribosome stalled on a poly(A) tail | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Chandrasekaran, V, Juszkiewicz, S, Choi, J, Puglisi, J.D, Brown, A, Shao, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2019-08-03 | | Release date: | 2019-12-04 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of ribosome stalling during translation of a poly(A) tail.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6SNE
 
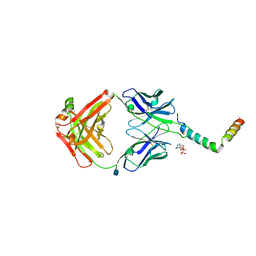 | | crystal structure of LN01 Fab in complex with an HIV-1 gp41 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, LN01 heavy chain, ... | | Authors: | Caillat, C, Pinto, D, Corti, D, Fenwick, C, Pantaleo, G, Weissenhorn, W. | | Deposit date: | 2019-08-23 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Basis for Broad HIV-1 Neutralization by the MPER-Specific Human Broadly Neutralizing Antibody LN01.
Cell Host Microbe, 26, 2019
|
|
6QWV
 
 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Isupov, N.M, Opatowsky, Y. | | Deposit date: | 2019-03-06 | | Release date: | 2019-07-03 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural Evidence for an Octameric Ring Arrangement of SARM1.
J.Mol.Biol., 431, 2019
|
|
4DGS
 
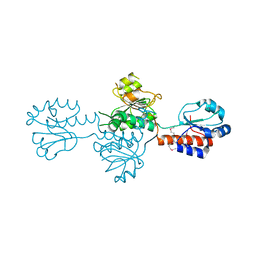 | | The crystals structure of dehydrogenase from Rhizobium meliloti | | Descriptor: | Dehydrogenase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-01-26 | | Release date: | 2012-02-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystals structure of dehydrogenase from Rhizobium meliloti
To be Published
|
|
6BTZ
 
 | | Crystal structure of the PI3KC2alpha C2 domain in space group C121 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, GLYCEROL, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, ... | | Authors: | Chen, K.-E, Collins, B.M. | | Deposit date: | 2017-12-08 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Basis for Membrane Recruitment by the PX and C2 Domains of Class II Phosphoinositide 3-Kinase-C2 alpha.
Structure, 26, 2018
|
|
6BST
 
 | |
4NF9
 
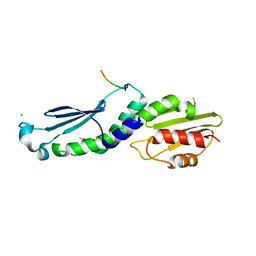 | | Structure of the Knl1/Nsl1 complex | | Descriptor: | CHLORIDE ION, Kinetochore-associated protein NSL1 homolog, Protein CASC5 | | Authors: | Petrovic, A, Mosalaganti, S, Keller, J, Mattiuzzo, M, Overlack, K, Wohlgemuth, S, Pasqualato, S, Raunser, S, Musacchio, A. | | Deposit date: | 2013-10-31 | | Release date: | 2014-03-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Modular Assembly of RWD Domains on the Mis12 Complex Underlies Outer Kinetochore Organization.
Mol.Cell, 53, 2014
|
|
6CCL
 
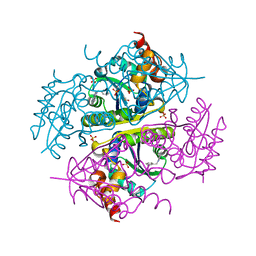 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 1-benzyl-1H-imidazo[4,5-b]pyridine | | Descriptor: | 1-benzyl-1H-imidazo[4,5-b]pyridine, DIMETHYL SULFOXIDE, Phosphopantetheine adenylyltransferase, ... | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria.
J. Med. Chem., 61, 2018
|
|
4NPQ
 
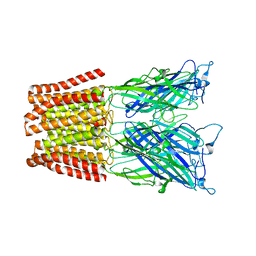 | | The resting-state conformation of the GLIC ligand-gated ion channel | | Descriptor: | Proton-gated ion channel | | Authors: | Sauguet, L, Shahsavar, A, Poitevin, F, Huon, C, Menny, A, Nemecz, A, Haouz, A, Changeux, J.P, Corringer, P.J, Delarue, M. | | Deposit date: | 2013-11-22 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6SND
 
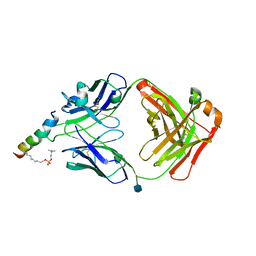 | | crystal structure of LN01 Fab in complex with an HIV-1 gp41 peptide | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Caillat, C, Pinto, D, Corti, D, Fenwick, C, Pantaleo, G, Weissenhorn, W. | | Deposit date: | 2019-08-23 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Broad HIV-1 Neutralization by the MPER-Specific Human Broadly Neutralizing Antibody LN01.
Cell Host Microbe, 26, 2019
|
|
6CNU
 
 | | Crystal Structure of JzTX-V | | Descriptor: | BROMIDE ION, GLYCEROL, JzTx-V, ... | | Authors: | Min, X, Wang, Z. | | Deposit date: | 2018-03-09 | | Release date: | 2019-03-06 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Discovery of Tarantula Venom-Derived NaV1.7-Inhibitory JzTx-V Peptide 5-Br-Trp24 Analogue AM-6120 with Systemic Block of Histamine-Induced Pruritis.
J. Med. Chem., 61, 2018
|
|
6CKA
 
 | | Crystal Structure of Paratox | | Descriptor: | Paratox | | Authors: | Prehna, G. | | Deposit date: | 2018-02-27 | | Release date: | 2018-11-14 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.559 Å) | | Cite: | The conserved mosaic prophage protein paratox inhibits the natural competence regulator ComR in Streptococcus.
Sci Rep, 8, 2018
|
|
6CUG
 
 | | Crystal structure of BC8B TCR-CD1b-PC complex | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Shahine, A.E, Rossjohn, J. | | Deposit date: | 2018-03-26 | | Release date: | 2019-01-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A T-cell receptor escape channel allows broad T-cell response to CD1b and membrane phospholipids.
Nat Commun, 10, 2019
|
|
6CUF
 
 | | Cryo-EM structure at 4.2 A resolution of vaccine-elicited antibody vFP1.01 in complex with HIV-1 Env BG505 DS-SOSIP, and antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Acharya, P, Carragher, B, Potter, C.S, Kwong, P.D. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Complete functional mapping of infection- and vaccine-elicited antibodies against the fusion peptide of HIV.
PLoS Pathog., 14, 2018
|
|
6SUA
 
 | |
6D2Q
 
 | | Crystal structure of the FERM domain of zebrafish FARP1 | | Descriptor: | FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) | | Authors: | Kuo, Y.C, Zhang, X. | | Deposit date: | 2018-04-13 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural analyses of FERM domain-mediated membrane localization of FARP1.
Sci Rep, 8, 2018
|
|
