5RZV
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z31504642 | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-N-phenylacetamide, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
8GXE
 
 | | PTPN21 FERM PTP complex | | Descriptor: | CHLORIDE ION, Tyrosine-protein phosphatase non-receptor type 21 | | Authors: | Chen, L, Zheng, Y.Y, Zhou, C. | | Deposit date: | 2022-09-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of PTPN21 reveals a dominant-negative effect of the FERM domain on its phosphatase activity.
Sci Adv, 10, 2024
|
|
5D68
 
 | | Crystal structure of KRIT1 ARD-FERM | | Descriptor: | Krev interaction trapped protein 1 | | Authors: | Zhang, R, Li, X, Boggon, T.J. | | Deposit date: | 2015-08-11 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structural analysis of the KRIT1 ankyrin repeat and FERM domains reveals a conformationally stable ARD-FERM interface.
J.Struct.Biol., 192, 2015
|
|
4HDO
 
 | | Crystal structure of the binary Complex of KRIT1 bound to the Rap1 GTPase | | Descriptor: | GLYCEROL, Krev interaction trapped protein 1, MAGNESIUM ION, ... | | Authors: | Gingras, A.R. | | Deposit date: | 2012-10-02 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Structure of the Ternary Complex of Krev Interaction Trapped 1 (KRIT1) Bound to Both the Rap1 GTPase and the Heart of Glass (HEG1) Cytoplasmic Tail.
J.Biol.Chem., 288, 2013
|
|
6OQ3
 
 | |
6OQ4
 
 | |
4HDQ
 
 | |
2YS3
 
 | | Solution structure of the PH domain of Kindlin-3 from human | | Descriptor: | Unc-112-related protein 2 | | Authors: | Li, H, Sato, M, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PH domain of Kindlin-3 from human
To be Published
|
|
7C3M
 
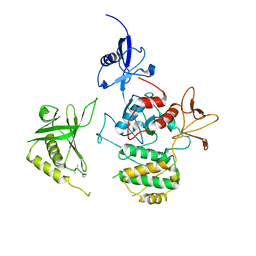 | | Structure of FERM protein | | Descriptor: | Fermitin family homolog 3,Fermitin family homolog 3,Fermitin family homolog 3 | | Authors: | Bu, W, Loh, Z.Y, Jin, S, Basu, S, Ero, R, Park, J.E, Yan, X, Wang, M, Sze, S.K, Tan, S.M, Gao, Y.G. | | Deposit date: | 2020-05-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis of human full-length kindlin-3 homotrimer in an auto-inhibited state.
Plos Biol., 18, 2020
|
|
6T36
 
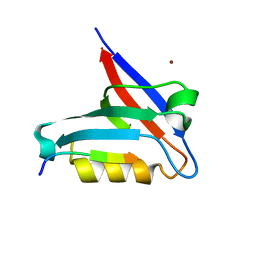 | | Crystal structure of the PTPN3 PDZ domain bound to the HBV core protein C-terminal peptide | | Descriptor: | BROMIDE ION, Capsid protein, Tyrosine-protein phosphatase non-receptor type 3 | | Authors: | Genera, M, Mechaly, A, Haouz, A, Caillet-Saguy, C. | | Deposit date: | 2019-10-10 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Molecular basis of the interaction of the human tyrosine phosphatase PTPN3 with the hepatitis B virus core protein.
Sci Rep, 11, 2021
|
|
5L81
 
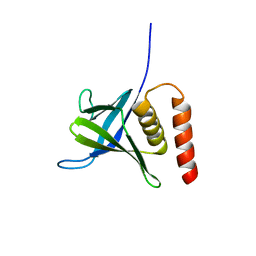 | |
4BBK
 
 | | Structural and functional characterisation of the kindlin-1 pleckstrin homology domain | | Descriptor: | FERMITIN FAMILY HOMOLOG 1, GLYCEROL | | Authors: | Yates, L.A, Lumb, C.N, Brahme, N.N, Zalyte, R, Bird, L.E, De Colibus, L, Owens, R.J, Calderwood, D.A, Sansom, M.S.P, Gilbert, R.J.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterisation of the Kindlin-1 Pleckstrin Homology Domain
J.Biol.Chem., 287, 2012
|
|
4DXA
 
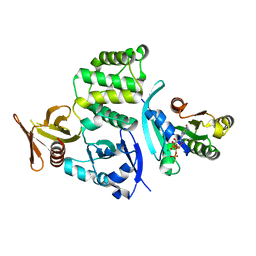 | | Co-crystal structure of Rap1 in complex with KRIT1 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Krev interaction trapped protein 1, MAGNESIUM ION, ... | | Authors: | Li, X, Zhang, R, Boggon, T.J. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for Small G Protein Effector Interaction of Ras-related Protein 1 (Rap1) and Adaptor Protein Krev Interaction Trapped 1 (KRIT1).
J.Biol.Chem., 287, 2012
|
|
4F7H
 
 | | The crystal structure of kindlin-2 pleckstrin homology domain in free form | | Descriptor: | Fermitin family homolog 2, S,R MESO-TARTARIC ACID | | Authors: | Liu, Y, Zhu, Y, Qin, J, Ye, S, Zhang, R. | | Deposit date: | 2012-05-16 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of kindlin-2 PH domain reveals a conformational transition for its membrane anchoring and regulation of integrin activation.
Protein Cell, 3, 2012
|
|
2LKO
 
 | | Structural Basis of Phosphoinositide Binding to Kindlin-2 Pleckstrin Homology Domain in Regulating Integrin Activation | | Descriptor: | Fermitin family homolog 2, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE | | Authors: | Liu, J, Fukuda, K, Xu, Z. | | Deposit date: | 2011-10-17 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of phosphoinositide binding to kindlin-2 protein pleckstrin homology domain in regulating integrin activation.
J.Biol.Chem., 286, 2011
|
|
6UZK
 
 | |
8SU8
 
 | |
8T7V
 
 | |
8T09
 
 | |
