6NUC
 
 | | Structure of Calcineurin in complex with NHE1 peptide | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, X, Page, R, Peti, W. | | Deposit date: | 2019-01-31 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for the binding and selective dephosphorylation of Na+/H+exchanger 1 by calcineurin.
Nat Commun, 10, 2019
|
|
6NTS
 
 | | Protein Phosphatase 2A (Aalpha-B56alpha-Calpha) holoenzyme in complex with a Small Molecule Activator of PP2A (SMAP) | | Descriptor: | MANGANESE (II) ION, N-[(1R,2R,3S)-2-hydroxy-3-(10H-phenoxazin-10-yl)cyclohexyl]-4-(trifluoromethoxy)benzene-1-sulfonamide, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform, ... | | Authors: | Huang, W, Taylor, D. | | Deposit date: | 2019-01-30 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Selective PP2A Enhancement through Biased Heterotrimer Stabilization.
Cell, 181, 2020
|
|
6NUU
 
 | | Structure of Calcineurin mutant in complex with NHE1 peptide | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, X, Page, R, Peti, W. | | Deposit date: | 2019-02-02 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the binding and selective dephosphorylation of Na+/H+exchanger 1 by calcineurin.
Nat Commun, 10, 2019
|
|
6NUF
 
 | | Structure of Calcineurin in complex with NHE1 peptide | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, X, Page, R, Peti, W. | | Deposit date: | 2019-01-31 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for the binding and selective dephosphorylation of Na+/H+exchanger 1 by calcineurin.
Nat Commun, 10, 2019
|
|
1HP1
 
 | | 5'-NUCLEOTIDASE (OPEN FORM) COMPLEX WITH ATP | | Descriptor: | 5'-NUCLEOTIDASE, ADENOSINE-5'-TRIPHOSPHATE, CARBONATE ION, ... | | Authors: | Knoefel, T, Straeter, N. | | Deposit date: | 2000-12-12 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of hydrolysis of phosphate esters by the dimetal center of 5'-nucleotidase based on crystal structures.
J.Mol.Biol., 309, 2001
|
|
6ZEI
 
 | | Structure of PP1-IRSp53 S455E chimera [PP1(7-304) + linker (G/S)x9 + IRSp53(449-465)] bound to Phactr1 (516-580) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZEH
 
 | | Structure of PP1-spectrin alpha II chimera [PP1(7-304) + linker (G/S)x9 + spectrin alpha II (1025-1039)] bound to Phactr1 (516-580) | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Phosphatase and actin regulator, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6Z9D
 
 | |
6ZEE
 
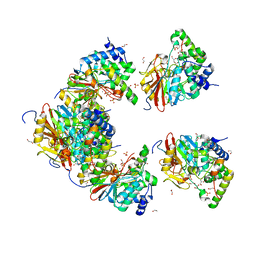 | | Structure of PP1(7-300) bound to Phactr1 (507-580) at pH8.4 | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE, GLYCEROL, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZEF
 
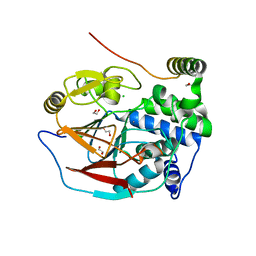 | | Structure of PP1(7-300) bound to Phactr1 (516-580) at pH 5.25 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZEG
 
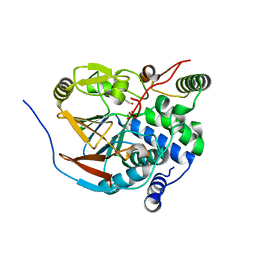 | | Structure of PP1-IRSp53 chimera [PP1(7-304) + linker (G/S)x9 + IRSp53(449-465)] bound to Phactr1 (516-580) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE, MANGANESE (II) ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZK6
 
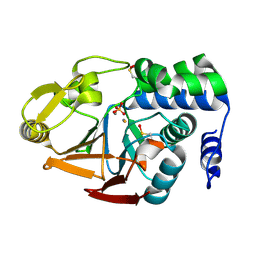 | | Protein Phosphatase 1 (PP1) T320E mutant | | Descriptor: | FE (III) ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Salvi, F, Barabas, O, Koehn, M. | | Deposit date: | 2020-06-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Towards Dissecting the Mechanism of Protein Phosphatase-1 Inhibition by Its C-Terminal Phosphorylation.
Chembiochem, 22, 2021
|
|
1II7
 
 | | Crystal structure of P. furiosus Mre11 with manganese and dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, Mre11 nuclease, ... | | Authors: | Hopfner, K.-P, Karcher, A, Craig, L, Woo, T.T, Carney, J.P, Tainer, J.A. | | Deposit date: | 2001-04-20 | | Release date: | 2001-05-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase.
Cell(Cambridge,Mass.), 105, 2001
|
|
1KBP
 
 | | KIDNEY BEAN PURPLE ACID PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, PURPLE ACID PHOSPHATASE, ... | | Authors: | Klabunde, T, Strater, N, Krebs, B. | | Deposit date: | 1995-02-20 | | Release date: | 1996-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Mechanism of Fe(III)-Zn(II) purple acid phosphatase based on crystal structures.
J.Mol.Biol., 259, 1996
|
|
1JK7
 
 | | CRYSTAL STRUCTURE OF THE TUMOR-PROMOTER OKADAIC ACID BOUND TO PROTEIN PHOSPHATASE-1 | | Descriptor: | BETA-MERCAPTOETHANOL, MANGANESE (II) ION, OKADAIC ACID, ... | | Authors: | Maynes, J.T, Bateman, K.S, Cherney, M.M, Das, A.K, James, M.N. | | Deposit date: | 2001-07-11 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the tumor-promoter okadaic acid bound to protein phosphatase-1.
J.Biol.Chem., 276, 2001
|
|
2BDX
 
 | | X-ray Crystal Structure of dihydromicrocystin-LA bound to Protein Phosphatase-1 | | Descriptor: | DIHYDROMICROCYSTIN-LA, MANGANESE (II) ION, Serine/threonine protein phosphatase PP1-gamma catalytic subunit | | Authors: | Maynes, J.T, Luu, H.A, Cherney, M.M, Andersen, R.J, Williams, D, Holmes, C.F, James, M.N. | | Deposit date: | 2005-10-21 | | Release date: | 2006-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Protein Phosphatase-1 Bound to Motuporin and Dihydromicrocystin-LA: Elucidation of the Mechanism of Enzyme Inhibition by Cyanobacterial Toxins.
J.Mol.Biol., 356, 2006
|
|
2BCD
 
 | | X-ray crystal structure of Protein Phosphatase-1 with the marine toxin motuporin bound | | Descriptor: | BETA-MERCAPTOETHANOL, MANGANESE (II) ION, MOTUPORIN, ... | | Authors: | Maynes, J.T, Luu, H.A, Cherney, M.M, Andersen, R.J, Williams, D, Holmes, C.F, James, M.N. | | Deposit date: | 2005-10-19 | | Release date: | 2006-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Protein Phosphatase-1 Bound to Motuporin and Dihydromicrocystin-LA: Elucidation of the Mechanism of Enzyme Inhibition by Cyanobacterial Toxins.
J.Mol.Biol., 356, 2006
|
|
1IT6
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN CALYCULIN A AND THE CATALYTIC SUBUNIT OF PROTEIN PHOSPHATASE 1 | | Descriptor: | CALYCULIN A, MANGANESE (II) ION, SERINE/THREONINE PROTEIN PHOSPHATASE 1 GAMMA (PP1-GAMMA) CATALYTIC SUBUNIT | | Authors: | Kita, A, Matsunaga, S, Takai, A, Kataiwa, H, Wakimoto, T, Fusetani, N, Isobe, M, Miki, K. | | Deposit date: | 2002-01-09 | | Release date: | 2002-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex between calyculin A and the catalytic subunit of protein phosphatase 1.
Structure, 10, 2002
|
|
1M63
 
 | | Crystal structure of calcineurin-cyclophilin-cyclosporin shows common but distinct recognition of immunophilin-drug complexes | | Descriptor: | CALCINEURIN B SUBUNIT ISOFORM 1, CALCIUM ION, CYCLOSPORIN A, ... | | Authors: | Huai, Q, Kim, H.-Y, Liu, Y, Zhao, Y, Mondragon, A, Liu, J.O, Ke, H. | | Deposit date: | 2002-07-12 | | Release date: | 2002-09-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Calcineurin-Cyclophilin-Cyclosporin Shows Common But Distinct Recognition of Immunophilin-Drug Complexes
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MF8
 
 | |
7YZP
 
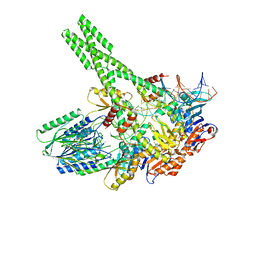 | | Hairpin-bound state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and a DNA hairpin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA hairpin (59-MER), MAGNESIUM ION, ... | | Authors: | Gut, F, Kaeshammer, L, Lammens, K, Bartho, J, van de Logt, E, Kessler, B, Hopfner, K.P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-17 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50.
Mol.Cell, 82, 2022
|
|
7Z03
 
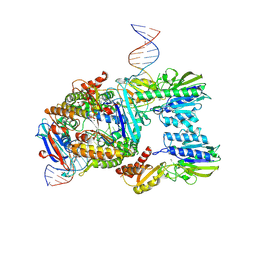 | | Endonuclease state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and extended dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (39-MER), MAGNESIUM ION, ... | | Authors: | Gut, F, Kaeshammer, L, Lammens, K, Bartho, J, van de Logt, E, Kessler, B, Hopfner, K.P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-17 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50.
Mol.Cell, 82, 2022
|
|
7YZO
 
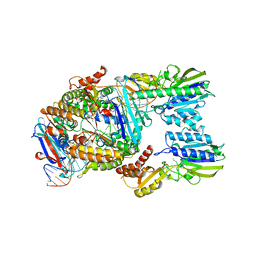 | | Endonuclease state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (31-MER), MAGNESIUM ION, ... | | Authors: | Gut, F, Kaeshammer, L, Lammens, K, Bartho, J, van de Logt, E, Kessler, B, Hopfner, K.P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-17 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50.
Mol.Cell, 82, 2022
|
|
2IE4
 
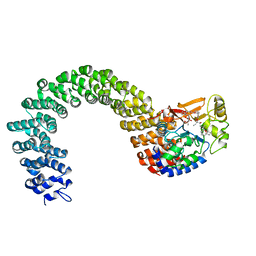 | | Structure of the Protein Phosphatase 2A Core Enzyme Bound to okadaic acid | | Descriptor: | MANGANESE (II) ION, OKADAIC ACID, Protein Phosphatase 2, ... | | Authors: | Xing, Y, Xu, Y, Chen, Y, Jeffrey, P.D, Chao, Y, Shi, Y. | | Deposit date: | 2006-09-17 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Protein Phosphatase 2A Core Enzyme Bound to Tumor-Inducing Toxins
Cell(Cambridge,Mass.), 127, 2006
|
|
2IAE
 
 | | Crystal structure of a protein phosphatase 2A (PP2A) holoenzyme. | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Cho, U.S, Xu, W. | | Deposit date: | 2006-09-07 | | Release date: | 2006-12-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme.
Nature, 445, 2007
|
|
