4MYQ
 
 | |
5MT2
 
 | | Glycoside hydrolase BT_0996 | | Descriptor: | Beta-galactosidase, GLYCEROL | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
4MXF
 
 | |
4MY2
 
 | | Crystal Structure of Norrin in fusion with Maltose Binding Protein | | Descriptor: | Maltose-binding periplasmic protein, Norrin fusion protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ke, J, Jurecky, C, Chen, C, Gu, X, Parker, N, Williams, B.O, Melcher, K, Xu, H.E. | | Deposit date: | 2013-09-27 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and function of Norrin in assembly and activation of a Frizzled 4-Lrp5/6 complex.
Genes Dev., 27, 2013
|
|
4N0J
 
 | | Crystal structure of dimethyllysine hen egg-white lysozyme in complex with sclx4 at 1.9 A resolution | | Descriptor: | 25,26,27,28-tetrahydroxypentacyclo[19.3.1.1~3,7~.1~9,13~.1~15,19~]octacosa-1(25),3(28),4,6,9(27),10,12,15(26),16,18,21,23-dodecaene-5,11,17,23-tetrasulfonic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | McGovern, R.E, Crowley, P.B. | | Deposit date: | 2013-10-02 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural study of a small molecule receptor bound to dimethyllysine in lysozyme.
Chem Sci, 6, 2015
|
|
6JG6
 
 | | Crystal structure of barley exohydrolaseI W286A mutant in complex with methyl 6-thio-beta-gentiobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, GLYCEROL, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
4MYZ
 
 | |
5MUJ
 
 | | BT0996 RGII Chain B Complex | | Descriptor: | ACETATE ION, Beta-galactosidase, alpha-L-rhamnopyranose-(1-2)-[alpha-L-rhamnopyranose-(1-3)]alpha-L-arabinopyranose-(1-4)-[4-O-[(1R)-1-hydroxyethyl]-2-O-methyl-alpha-L-fucopyranose-(1-2)]beta-D-galactopyranose-(1-2)-alpha-D-aceric acid-(1-4)-alpha-L-rhamnopyranose-(1-3)-3-C-(hydroxylmethyl)-alpha-D-erythrofuranose | | Authors: | Cartmell, A, Basle, A, Ndeh, D, Luis, A.S, Venditto, I, Labourel, A, Rogowski, A, Gilbert, H.J. | | Deposit date: | 2017-01-13 | | Release date: | 2017-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
6JGT
 
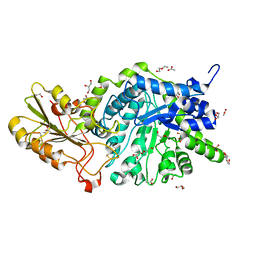 | | Crystal structure of barley exohydrolaseI W434Y mutant in complex with methyl 6-thio-beta-gentiobioside. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
4N1U
 
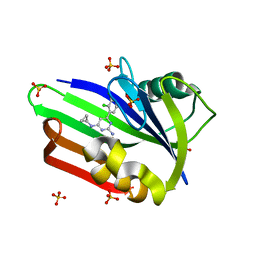 | | Structure of human MTH1 in complex with TH588 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N~4~-cyclopropyl-6-(2,3-dichlorophenyl)pyrimidine-2,4-diamine, SULFATE ION | | Authors: | Berntsson, R.P.-A, Jemth, A, Gustafsson, R, Svensson, L.M, Helleday, T, Stenmark, P. | | Deposit date: | 2013-10-04 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool.
Nature, 508, 2014
|
|
6JHW
 
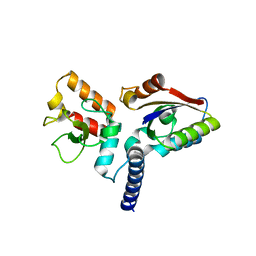 | | Structure of anti-CRISPR AcrIIC3 and NmeCas9 HNH | | Descriptor: | AcrIIC3, CRISPR-associated endonuclease Cas9 | | Authors: | Suh, J.Y, Lee, B.J, Lee, S.J, Kim, Y. | | Deposit date: | 2019-02-19 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Anti-CRISPR AcrIIC3 discriminates between Cas9 orthologs via targeting the variable surface of the HNH nuclease domain.
Febs J., 286, 2019
|
|
4N2Z
 
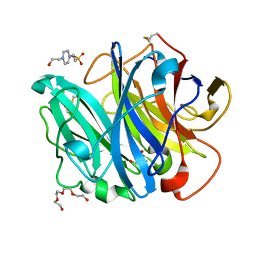 | | Crystal Structure of the alpha-L-arabinofuranosidase PaAbf62A from Podospora anserina in complex with cellotriose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Siguier, B, Dumon, C, Mourey, L, Tranier, S. | | Deposit date: | 2013-10-06 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | First Structural Insights into alpha-L-Arabinofuranosidases from the Two GH62 Glycoside Hydrolase Subfamilies.
J.Biol.Chem., 289, 2014
|
|
4N2X
 
 | | Crystal Structure of DL-2-haloacid dehalogenase | | Descriptor: | DL-2-haloacid dehalogenase, GLYCEROL | | Authors: | Siwek, A, Omi, R, Hirotsu, K, Jitsumori, K, Esaki, N, Kurihara, T, Paneth, P. | | Deposit date: | 2013-10-06 | | Release date: | 2013-11-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding modes of DL-2-haloacid dehalogenase revealed by crystallography, modeling and isotope effects studies.
Arch.Biochem.Biophys., 540, 2013
|
|
4N4J
 
 | | Kuenenia stuttgartiensis hydroxylamine oxidoreductase | | Descriptor: | 1-[(4-cyclohexylbutanoyl)(2-hydroxyethyl)amino]-1-deoxy-D-glucitol, HEME C, PHOSPHATE ION, ... | | Authors: | Maalcke, W.J, Dietl, A, Marritt, S.J, Butt, J.N, Jetten, M.S.M, Keltjens, J.T, Barends, T.R.M.B, Kartal, B. | | Deposit date: | 2013-10-08 | | Release date: | 2013-12-11 | | Last modified: | 2020-03-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Biological NO Generation by Octaheme Oxidoreductases.
J.Biol.Chem., 289, 2014
|
|
4N4S
 
 | | A Double Mutant Rat Erk2 in Complex With a Pyrazolo[3,4-d]pyrimidine Inhibitor | | Descriptor: | 3-[2-(benzyloxy)-8-methylquinolin-6-yl]-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Mitogen-activated protein kinase 1 | | Authors: | Hari, S.B, Maly, D.J, Merritt, E.A. | | Deposit date: | 2013-10-08 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformation-Selective ATP-Competitive Inhibitors Control Regulatory Interactions and Noncatalytic Functions of Mitogen-Activated Protein Kinases.
Chem.Biol., 21, 2014
|
|
4N7G
 
 | | Crystal structure of 14-3-3zeta in complex with a peptide derived from ExoS | | Descriptor: | 14-3-3 protein zeta/delta, Exoenzyme S, HEXAETHYLENE GLYCOL | | Authors: | Bier, D, Glas, A, Hahne, G, Grossmann, T, Ottmann, C. | | Deposit date: | 2013-10-15 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Constrained peptides with target-adapted cross-links as inhibitors of a pathogenic protein-protein interaction.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5TPC
 
 | | Binding domain of BoNT/A complexed with ganglioside | | Descriptor: | Botulinum neurotoxin type A, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose | | Authors: | Berntsson, R.P.-A, Svensson, L.M, Stenmark, P. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Glycans Confer Specificity to the Recognition of Ganglioside Receptors by Botulinum Neurotoxin A.
J. Am. Chem. Soc., 139, 2017
|
|
4N8S
 
 | | Crystal Structure of the ternary complex of lipase from Thermomyces lanuginosa with Ethylacetoacetate and P-nitrobenzaldehyde at 2.3 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-nitrobenzaldehyde, GLYCEROL, ... | | Authors: | Kumar, M, Mukherjee, J, Gupta, M.N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-10-18 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the ternary complex of lipase from Thermomyces lanuginosa with Ethylacetoacetate and P-nitrobenzaldehyde at 2.3 A resolution
To be Published
|
|
6JOM
 
 | | Crystal structure of lipoate protein ligase from Mycoplasma hyopneumoniae | | Descriptor: | 5'-O-[(R)-({5-[(3R)-1,2-DITHIOLAN-3-YL]PENTANOYL}OXY)(HYDROXY)PHOSPHORYL]ADENOSINE, Lipoate--protein ligase | | Authors: | Zhang, H, Chen, H, Ma, G. | | Deposit date: | 2019-03-22 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional Identification and Structural Analysis of a New Lipoate Protein Ligase inMycoplasma hyopneumoniae.
Front Cell Infect Microbiol, 10, 2020
|
|
4NA4
 
 | | Crystal structure of mouse poly(ADP-ribose) glycohydrolase (PARG) catalytic domain with ADP-HPD | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, IODIDE ION, Poly(ADP-ribose) glycohydrolase | | Authors: | Wang, Z, Cheng, Z, Xu, W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic and biochemical analysis of the mouse poly(ADP-ribose) glycohydrolase.
Plos One, 9, 2014
|
|
4NB1
 
 | | Crystal Structure of FosB from Staphylococcus aureus at 1.80 Angstrom Resolution with L-Cysteine-Cys9 Disulfide | | Descriptor: | CYSTEINE, Metallothiol transferase FosB, SULFATE ION | | Authors: | Cook, P.D, Thompson, M.K, Goodman, M.C, Jagessar, K, Harp, J, Keithly, M.E, Armstrong, R.N. | | Deposit date: | 2013-10-22 | | Release date: | 2014-02-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Function of the Genomically Encoded Fosfomycin Resistance Enzyme, FosB, from Staphylococcus aureus.
Biochemistry, 53, 2014
|
|
6JLH
 
 | | Structure of SCGN in complex with a Snap25 peptide | | Descriptor: | CALCIUM ION, CHLORIDE ION, Secretagogin, ... | | Authors: | Qin, J, Sun, Q, Jia, D. | | Deposit date: | 2019-03-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural and mechanistic insights into secretagogin-mediated exocytosis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4N82
 
 | | X-ray crystal structure of Streptococcus sanguinis NrdIox | | Descriptor: | FLAVIN MONONUCLEOTIDE, Ribonucleotide reductase, SULFATE ION | | Authors: | Boal, A.K, Rosenzweig, A.C. | | Deposit date: | 2013-10-16 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Streptococcus sanguinis Class Ib Ribonucleotide Reductase: HIGH ACTIVITY WITH BOTH IRON AND MANGANESE COFACTORS AND STRUCTURAL INSIGHTS.
J.Biol.Chem., 289, 2014
|
|
5MAG
 
 | | Crystal structure of MELK in complex with an inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Maternal embryonic leucine zipper kinase, ... | | Authors: | Canevari, G, Re Depaolini, S, Casale, E, Felder, E, Kuster, B, Heinzlmeir, S. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
5MGD
 
 | | STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 6-Galactosyl-lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Rico-Diaz, A, Ramirez-Escudero, M, Vizoso Vazquez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2016-11-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural features of Aspergillus niger beta-galactosidase define its activity against glycoside linkages.
FEBS J., 284, 2017
|
|
