6HNJ
 
 | | The ligand-bound, open structure of CD0873, a substrate binding protein with adhesive properties from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, ABC-type transport system, sugar-family extracellular solute-binding protein, ... | | Authors: | Bradshaw, W.J, Kovacs-Simon, A, Harmer, N.J, Michell, S.L, Acharya, K.R. | | Deposit date: | 2018-09-15 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular features of lipoprotein CD0873: A potential vaccine against the human pathogenClostridioides difficile.
J.Biol.Chem., 294, 2019
|
|
2WYJ
 
 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with monoaryl P4 motifs | | Descriptor: | (E)-2-(5-CHLOROTHIOPHEN-2-YL)-N-[(3S)-1-{4-[(1S)-1-(DIMETHYLAMINO)ETHYL]-2-FLUOROPHENYL}-2-OXOPYRROLIDIN-3-YL]ETHENESULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, FACTOR X LIGHT CHAIN | | Authors: | Kleanthous, S, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chaudry, L, Chan, C, Clarte, M, Convery, M.A, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Pateman, A.J, Patikis, A, Pinto, I.L, Pollard, D.R, Roethka, T.J, Senger, S, Shah, G.P, Stelman, G.J, Toomey, J.R, Watson, N.S, Whittaker, C, Zhou, P, Young, R.J. | | Deposit date: | 2009-11-16 | | Release date: | 2010-12-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Monoaryl P4 Motifs
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5NS9
 
 | |
2G9J
 
 | | Complex of TM1a(1-14)Zip with TM9a(251-284): a model for the polymerization domain ("overlap region") of tropomyosin, Northeast Structural Genomics Target OR9 | | Descriptor: | Tropomyosin 1 alpha chain, Tropomyosin 1 alpha chain/General control protein GCN4 | | Authors: | Greenfield, N.J, Huang, Y.J, Swapna, G.V.T, Bhattacharya, A, Singh, A, Montelione, G.T, Hitchcock-DeGregori, S.E, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-03-06 | | Release date: | 2006-11-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Junction between Tropomyosin Molecules: Implications for Actin Binding and Regulation.
J.Mol.Biol., 364, 2006
|
|
4E9L
 
 | | FdeC, a Novel Broadly Conserved Escherichia coli Adhesin Eliciting Protection against Urinary Tract Infections | | Descriptor: | Attaching and effacing protein, pathogenesis factor | | Authors: | Spraggon, G, Nesta, B, Alteri, C, Gomes Moriel, D, Rosini, R, Veggi, D, Smith, S, Bertoldi, I, Pastorello, I, Ferlenghi, I, Fontana, M.R, Frankel, G, Mobley, H.L.T, Rappuoli, R, Pizza, M, Serino, L, Soriana, M. | | Deposit date: | 2012-03-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | FdeC, a novel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections.
MBio, 3, 2012
|
|
4OWL
 
 | |
1FAZ
 
 | |
2GV6
 
 | | Crystal Structure of Matriptase with Inhibitor CJ-730 | | Descriptor: | (S)-3-(3-(4-(2-GUANIDINOETHYL)PIPERIDIN-1-YL)-2-(NAPHTHALENE-2-SULFONAMIDO)-3-OXOPROPYL)BENZIMIDAMIDE, Suppressor of tumorigenicity 14 | | Authors: | Bode, W. | | Deposit date: | 2006-05-02 | | Release date: | 2006-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Secondary Amides of Sulfonylated 3-Amidinophenylalanine. New Potent and Selective Inhibitors of Matriptase.
J.Med.Chem., 49, 2006
|
|
4DHL
 
 | | Crystal structure of red kidney bean purple acid phosphatase in complex with Maybridge fragment MO07123 | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-methylphenyl)-1,3-thiazole-4-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feder, D, Clayton, D.J, Hussein, W.M, Schenk, G, McGeary, R, Guddat, L.W. | | Deposit date: | 2012-01-29 | | Release date: | 2012-12-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of purple acid phosphatase inhibitors by fragment-based screening: promising new leads for osteoporosis therapeutics.
Chem.Biol.Drug Des., 80, 2012
|
|
1DOJ
 
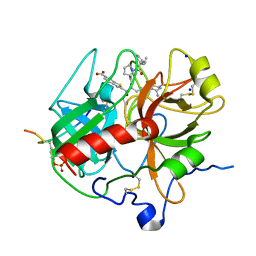 | | Crystal structure of human alpha-thrombin*RWJ-51438 complex at 1.7 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN, HIRUGEN, ... | | Authors: | Recacha, R, Costanzo, M.J, Maryanoff, B.E, Carson, M, DeLucas, L, Chattopadhyay, D. | | Deposit date: | 1999-12-21 | | Release date: | 2000-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human alpha-thrombin complexed with RWJ-51438 at 1.7 A: unusual perturbation of the 60A-60I insertion loop.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
4OWJ
 
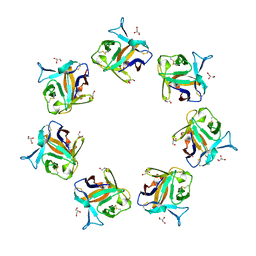 | |
2GV7
 
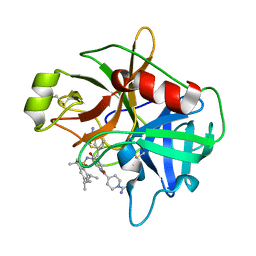 | | Structure of Matriptase in Complex with Inhibitor CJ-672 | | Descriptor: | (S)-4-(4-(3-(3-CARBAMIMIDOYLPHENYL)-2-(2,4,6-TRIISOPROPYLPHENYLSULFONAMIDO)PROPANOYL)PIPERAZINE-1-CARBONYL)PIPERIDINE-1-CARBOXIMIDAMIDE, Suppressor of tumorigenicity 14 | | Authors: | Bode, W. | | Deposit date: | 2006-05-02 | | Release date: | 2006-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Secondary Amides of Sulfonylated 3-Amidinophenylalanine. New Potent and Selective Inhibitors of Matriptase.
J.Med.Chem., 49, 2006
|
|
3PIQ
 
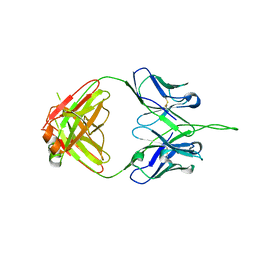 | | Crystal structure of human 2909 Fab, a quaternary structure-specific antibody against HIV-1 | | Descriptor: | Human monoclonal antibody 2909 Fab heavy chain, Human monoclonal antibody 2909 Fab light chain | | Authors: | Changela, A, Gorny, M.K, Zolla-Pazner, S, Kwong, P.D. | | Deposit date: | 2010-11-07 | | Release date: | 2011-01-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.325 Å) | | Cite: | Crystal Structure of Human Antibody 2909 Reveals Conserved Features of Quaternary Structure-Specific Antibodies That Potently Neutralize HIV-1.
J.Virol., 85, 2011
|
|
4EO8
 
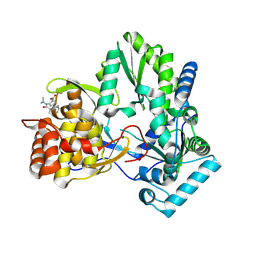 | | HCV NS5B polymerase inhibitors: Tri-substituted acylhydrazines as tertiary amide bioisosteres | | Descriptor: | 5-(3,3-dimethylbut-1-yn-1-yl)-3-{2,2-dimethyl-1-[(trans-4-methylcyclohexyl)carbonyl]hydrazinyl}thiophene-2-carboxylic acid, RNA-directed RNA polymerase | | Authors: | Appleby, T.C, Canales, E, Watkins, W.J. | | Deposit date: | 2012-04-13 | | Release date: | 2012-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Tri-substituted acylhydrazines as tertiary amide bioisosteres: HCV NS5B polymerase inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4HRT
 
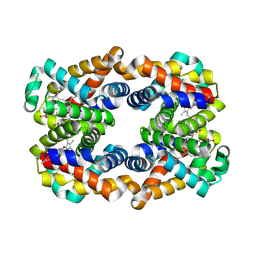 | | Scapharca tetrameric hemoglobin, unliganded | | Descriptor: | Globin-2 A chain, Hemoglobin B chain, PHOSPHATE ION, ... | | Authors: | Royer, W.E. | | Deposit date: | 2012-10-28 | | Release date: | 2012-12-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Tertiary and Quaternary Allostery in Tetrameric Hemoglobin from Scapharca inaequivalvis.
Biochemistry, 52, 2013
|
|
4HF4
 
 | |
2KPD
 
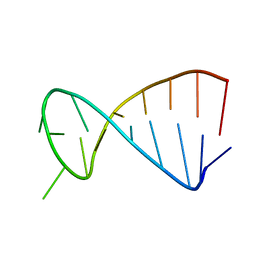 | | Structure determination of the top-loop of the conserved 3 terminal secondary structure in the genome of YFV-mutant | | Descriptor: | RNA (5'-R(*UP*GP*AP*GP*CP*UP*CP*AP*GP*UP*UP*UP*GP*CP*UP*CP*A)-3') | | Authors: | Lescrinier, E, Dyubankova, N, Nauwelaerts, K, Jones, R, Herdewijn, P. | | Deposit date: | 2009-10-12 | | Release date: | 2010-06-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure Determination of the Top-Loop of the Conserved 3'-Terminal Secondary Structure in the Genome of Flaviviruses.
Chembiochem, 11, 2010
|
|
4HEU
 
 | |
1I8F
 
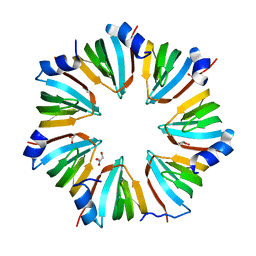 | | THE CRYSTAL STRUCTURE OF A HEPTAMERIC ARCHAEAL SM PROTEIN: IMPLICATIONS FOR THE EUKARYOTIC SNRNP CORE | | Descriptor: | GLYCEROL, PUTATIVE SNRNP SM-LIKE PROTEIN | | Authors: | Mura, C, Cascio, D, Sawaya, M.R, Eisenberg, D. | | Deposit date: | 2001-03-14 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of a heptameric archaeal Sm protein: Implications for the eukaryotic snRNP core.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2VTF
 
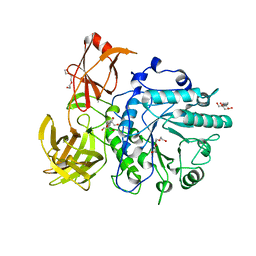 | | X-ray crystal structure of the Endo-beta-N-acetylglucosaminidase from Arthrobacter protophormiae E173Q mutant reveals a TIM barrel catalytic domain and two ancillary domains | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ENDO-BETA-N-ACETYLGLUCOSAMINIDASE, TRIETHYLENE GLYCOL | | Authors: | Ling, Z, Bingham, R.J, Suits, M.D.L, Moir, J.W.B, Fairbanks, A.J, Taylor, E.J. | | Deposit date: | 2008-05-14 | | Release date: | 2009-03-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The X-Ray Crystal Structure of an Arthrobacter Protophormiae Endo-Beta-N-Acetylglucosaminidase Reveals a (Beta/Alpha)(8) Catalytic Domain, Two Ancillary Domains and Active Site Residues Key for Transglycosylation Activity.
J.Mol.Biol., 389, 2009
|
|
3NV3
 
 | |
3O65
 
 | | Crystal structure of a Josephin-ubiquitin complex: Evolutionary restraints on ataxin-3 deubiquitinating activity | | Descriptor: | Putative ataxin-3-like protein, SODIUM ION, Ubiquitin | | Authors: | Weeks, S.D, Grasty, K.C, Hernandez-Cuebas, L, Loll, P.J. | | Deposit date: | 2010-07-28 | | Release date: | 2010-11-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of a Josephin-Ubiquitin Complex: EVOLUTIONARY RESTRAINTS ON ATAXIN-3 DEUBIQUITINATING ACTIVITY.
J.Biol.Chem., 286, 2011
|
|
2XAJ
 
 | | Crystal structure of LSD1-CoREST in complex with (-)-trans-2- phenylcyclopropyl-1-amine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, PHENYLETHYLENECARBOXYLIC ACID, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
4IKT
 
 | | Crystal structure of truncated (delta 1-89) human methionine aminopeptidase Type 1 in complex with N1-(5-chloro-6-methyl-2-(pyridin-2-yl)pyrimidin-4-yl)-N2-(5-(trifluoromethyl)pyridin-2-yl)ethane-1,2-diamine | | Descriptor: | COBALT (II) ION, GLYCEROL, Methionine aminopeptidase 1, ... | | Authors: | Addlagatta, A, Kishor, C, Arya, T. | | Deposit date: | 2012-12-28 | | Release date: | 2013-12-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification, Biochemical and Structural Evaluation of Species-Specific Inhibitors against Type I Methionine Aminopeptidases
J.Med.Chem., 56, 2013
|
|
3HNS
 
 | | CS-35 Fab Complex with Oligoarabinofuranosyl Hexasaccharide | | Descriptor: | CS-35 Fab Heavy Chain, CS-35 Fab Light Chain, beta-D-arabinofuranose-(1-2)-alpha-D-arabinofuranose-(1-3)-[beta-D-arabinofuranose-(1-2)-alpha-D-arabinofuranose-(1-5)]alpha-D-arabinofuranose-(1-5)-methyl alpha-D-arabinofuranoside | | Authors: | Murase, T, Zheng, R.B, Joe, M, Bai, Y, Marcus, S.L, Lowary, T.L, Ng, K.K.S. | | Deposit date: | 2009-06-01 | | Release date: | 2009-07-21 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into antibody recognition of mycobacterial polysaccharides.
J.Mol.Biol., 392, 2009
|
|
