6DG8
 
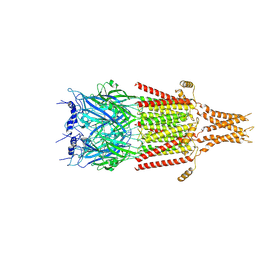 | | Full-length 5-HT3A receptor in a serotonin-bound conformation- State 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, SEROTONIN, ... | | Authors: | Basak, S, Chakrapani, S. | | Deposit date: | 2018-05-17 | | Release date: | 2018-11-07 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Cryo-EM reveals two distinct serotonin-bound conformations of full-length 5-HT3Areceptor.
Nature, 563, 2018
|
|
6DRV
 
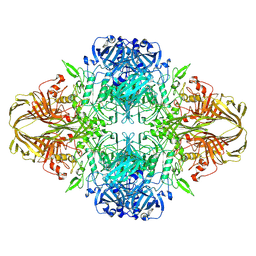 | |
6DHH
 
 | | RT XFEL structure of Photosystem II 400 microseconds after the second illumination at 2.2 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
3B8B
 
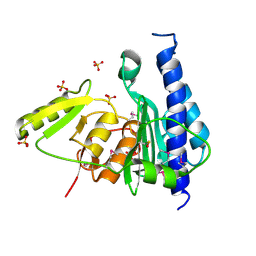 | | Crystal structure of CysQ from Bacteroides thetaiotaomicron, a bacterial member of the inositol monophosphatase family | | Descriptor: | CHLORIDE ION, CysQ, sulfite synthesis pathway protein, ... | | Authors: | Cuff, M.E, Mulligan, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-10-31 | | Release date: | 2007-12-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of CysQ from Bacteroides thetaiotaomicron, a bacterial member of the inositol monophosphatase family.
TO BE PUBLISHED
|
|
3BI3
 
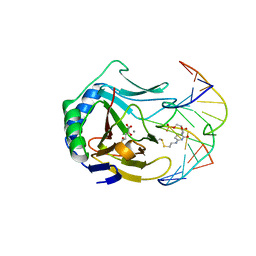 | |
3BCF
 
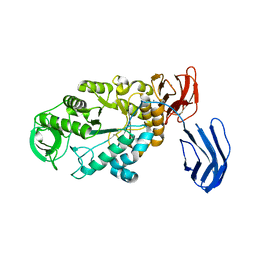 | | Alpha-amylase B from Halothermothrix orenii | | Descriptor: | Alpha amylase, catalytic region, CALCIUM ION, ... | | Authors: | Tan, T.-C, Mijts, B.N, Swaminathan, K, Patel, B.K.C, Divne, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Polyextremophilic alpha-Amylase AmyB from Halothermothrix orenii: Details of a Productive Enzyme-Substrate Complex and an N Domain with a Role in Binding Raw Starch
J.Mol.Biol., 378, 2008
|
|
3FMS
 
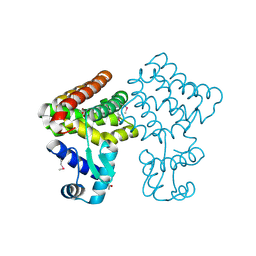 | | Crystal structure of TM0439, a GntR transcriptional regulator | | Descriptor: | ACETATE ION, NICKEL (II) ION, Transcriptional regulator, ... | | Authors: | Zheng, M, Cooper, D.R, Yu, M, Hung, L.-W, Derewenda, U, Derewenda, Z.S, Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2008-12-22 | | Release date: | 2009-02-10 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Thermotoga maritima TM0439: implications for the mechanism of bacterial GntR transcription regulators with Zn2+-binding FCD domains.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3FN1
 
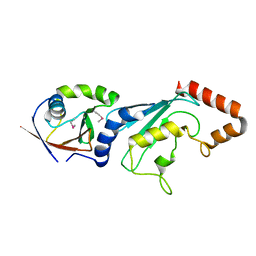 | | E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. | | Descriptor: | NEDD8-activating enzyme E1 catalytic subunit, NEDD8-conjugating enzyme UBE2F | | Authors: | Huang, D.T, Ayrault, O, Hunt, H.W, Taherbhoy, A.M, Duda, D.M, Scott, D.C, Borg, L.A, Neale, G, Murray, P.J, Roussel, M.F, Schulman, B.A. | | Deposit date: | 2008-12-22 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification
Mol.Cell, 33, 2009
|
|
3FB7
 
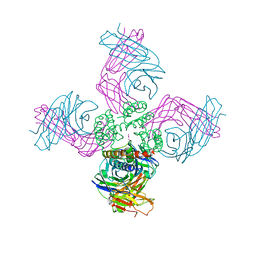 | | Open KcsA potassium channel in the presence of Rb+ ion | | Descriptor: | RUBIDIUM ION, Voltage-gated potassium channel, antibody fab fragment heavy chain, ... | | Authors: | Cuello, L.G, Jogini, V, Cortes, D.M, Pan, A.C, Gagnon, D.H, Cordero-Morales, J.F, Chakrapani, S, Roux, B, Perozo, E. | | Deposit date: | 2008-11-18 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Open KcsA potassium channel in the presence of Rb+ ion
TO BE PUBLISHED
|
|
3FH5
 
 | |
3F5K
 
 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3EXU
 
 | | A glycoside hydrolase family 11 xylanase with an extended thumb region | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase, GLYCEROL | | Authors: | Vandermarliere, E, Pollet, A, Strelkov, S.V, Delcour, J.A, Courtin, C.M. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystallographic and activity-based evidence for thumb flexibility and its relevance in glycoside hydrolase family 11 xylanases
Proteins, 77, 2009
|
|
3FBW
 
 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase DhaA mutant C176Y | | Descriptor: | BENZOIC ACID, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Dohnalek, J, Stsiapanava, A, Gavira, J.A, Kuta Smatanova, I, Kuty, M. | | Deposit date: | 2008-11-20 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Atomic resolution studies of haloalkane dehalogenases DhaA04, DhaA14 and DhaA15 with engineered access tunnels.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3DLA
 
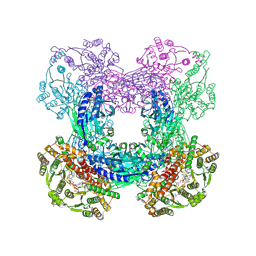 | | X-ray crystal structure of glutamine-dependent NAD+ synthetase from Mycobacterium tuberculosis bound to NaAD+ and DON | | Descriptor: | 5-OXO-L-NORLEUCINE, GLYCEROL, Glutamine-dependent NAD(+) synthetase, ... | | Authors: | LaRonde-LeBlanc, N.A, Resto, M, Gerratana, B. | | Deposit date: | 2008-06-26 | | Release date: | 2009-03-10 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Regulation of active site coupling in glutamine-dependent NAD(+) synthetase.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3D9D
 
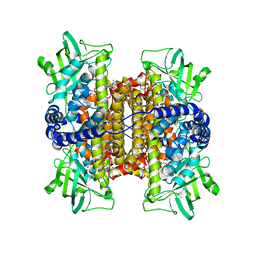 | | Nitroalkane oxidase: mutant D402N crystallized with 1-nitrohexane | | Descriptor: | 1-nitrohexane, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Heroux, A, Bozinovski, D.M, Valley, M.P, Fitzpatrick, P.F, Orville, A.M. | | Deposit date: | 2008-05-27 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of intermediates in the nitroalkane oxidase reaction.
Biochemistry, 48, 2009
|
|
3DRK
 
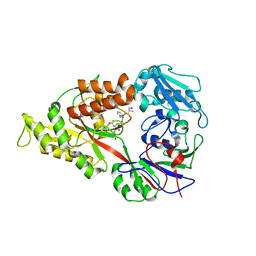 | | Crystal structure of Lactococcal OppA co-crystallized with Neuropeptide S in an open conformation | | Descriptor: | Neuropeptide S, Oligopeptide-binding protein oppA | | Authors: | Berntsson, R.P.-A, Doeven, M.K, Duurkens, R.H, Sengupta, D, Marrink, S.-J, Thunnissen, A.-M, Poolman, B, Slotboom, D.-J. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis for peptide selection by the transport receptor OppA
Embo J., 28, 2009
|
|
3DUQ
 
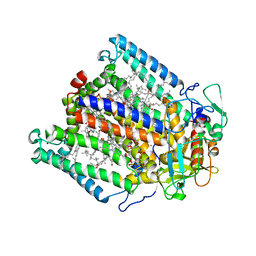 | |
3DKK
 
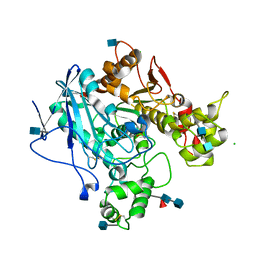 | | Aged Form of Human Butyrylcholinesterase Inhibited by Tabun | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nachon, F, Carletti, E. | | Deposit date: | 2008-06-25 | | Release date: | 2008-12-02 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Aging of Cholinesterases Phosphylated by Tabun Proceeds through O-Dealkylation.
J.Am.Chem.Soc., 130, 2008
|
|
3D85
 
 | | Crystal structure of IL-23 in complex with neutralizing FAB | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FAB of antibody 7G10, heavy chain, ... | | Authors: | Beyer, B.M, Ingram, R, Ramanathan, L, Reichert, P, Le, H, Madison, V. | | Deposit date: | 2008-05-22 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the pro-inflammatory cytokine interleukin-23 and its complex with a high-affinity neutralizing antibody
J.Mol.Biol., 382, 2008
|
|
3DL8
 
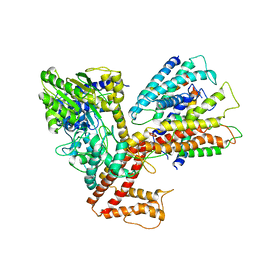 | | Structure of the complex of aquifex aeolicus SecYEG and bacillus subtilis SecA | | Descriptor: | Preprotein translocase subunit secY, Protein translocase subunit secA, Protein-export membrane protein secG, ... | | Authors: | Nam, Y, Zimmer, J, Rapoport, T.A. | | Deposit date: | 2008-06-26 | | Release date: | 2008-12-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (7.5 Å) | | Cite: | Structure of a complex of the ATPase SecA and the protein-translocation channel.
Nature, 455, 2008
|
|
3DOK
 
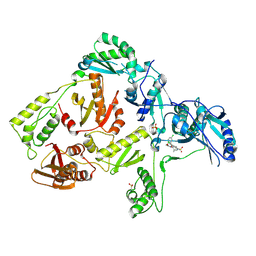 | | Crystal structure of K103N mutant HIV-1 reverse transcriptase in complex with GW678248. | | Descriptor: | 2-{4-chloro-2-[(3-chloro-5-cyanophenyl)carbonyl]phenoxy}-N-(2-methyl-4-sulfamoylphenyl)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Chamberlain, P.P, Ren, J, Stammers, D.K. | | Deposit date: | 2008-07-04 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
3DET
 
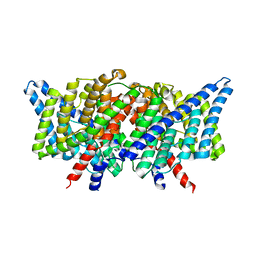 | | Structure of the E148A, Y445A doubly ungated mutant of E.coli CLC_Ec1, Cl-/H+ antiporter | | Descriptor: | Fab fragment, Heavy chain, Light chain, ... | | Authors: | Jayaram, H, Accardi, A, Wu, F, Williams, C, Miller, C. | | Deposit date: | 2008-06-10 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ion permeation through a Cl--selective channel designed from a CLC Cl-/H+ exchanger
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3DIN
 
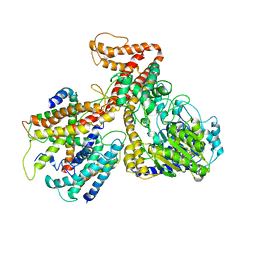 | | Crystal structure of the protein-translocation complex formed by the SecY channel and the SecA ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Zimmer, J, Nam, Y, Rapoport, T.A. | | Deposit date: | 2008-06-20 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structure of a complex of the ATPase SecA and the protein-translocation channel.
Nature, 455, 2008
|
|
3DLK
 
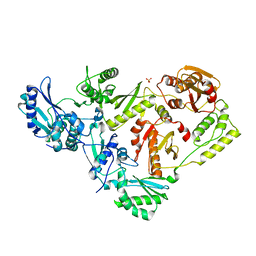 | | Crystal Structure of an engineered form of the HIV-1 Reverse Transcriptase, RT69A | | Descriptor: | Reverse transcriptase/ribonuclease H, SULFATE ION, p51 RT | | Authors: | Ho, W.C, Bauman, J.D, Himmel, D.M, Das, K, Arnold, E. | | Deposit date: | 2008-06-27 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal engineering of HIV-1 reverse transcriptase for structure-based drug design.
Nucleic Acids Res., 36, 2008
|
|
3DMJ
 
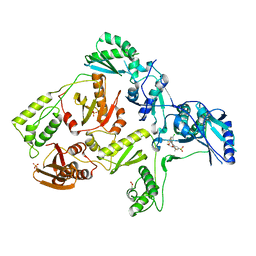 | | CRYSTAL STRUCTURE OF HIV-1 V106A and Y181C MUTANT REVERSE TRANSCRIPTASE IN COMPLEX WITH GW564511. | | Descriptor: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2008-07-01 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
