3QEU
 
 | | The crystal structure of TCR DMF5 | | Descriptor: | DMF5 alpha chain, DMF5 beta chain, GLYCEROL, ... | | Authors: | Borbulevych, O.Y, Santhanagopolan, S.M, Baker, B.M. | | Deposit date: | 2011-01-20 | | Release date: | 2011-07-06 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | TCRs Used in Cancer Gene Therapy Cross-React with MART-1/Melan-A Tumor Antigens via Distinct Mechanisms.
J.Immunol., 187, 2011
|
|
3Q8F
 
 | | Crystal structure of 2-Fluorohistine labeled Protective Antigen (pH 5.8) | | Descriptor: | CALCIUM ION, Protective antigen, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Battaile, K.P, Rajapaksha, M, Janowiak, B.E, Andra, K.K, Bann, J.G. | | Deposit date: | 2011-01-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | pH effects on binding between the anthrax protective antigen and the host cellular receptor CMG2.
Protein Sci., 21, 2012
|
|
3QF9
 
 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a furan-thiazolidinedione ligand | | Descriptor: | 6-{5-[(Z)-(2,4-dioxo-1,3-thiazolidin-5-ylidene)methyl]furan-2-yl}-N-{3-[(4-ethylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl}naphthalene-1-carboxamide, CHLORIDE ION, Proto-oncogene serine/threonine-protein kinase pim-1, ... | | Authors: | Filippakopoulos, P, Bullock, A.N, Fedorov, O, Miduturu, C.V, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Grey, N, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-21 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a furan-thiazolidinedione ligand
To be Published
|
|
3Q9F
 
 | | Crystal Structure of APAH complexed with CAPS | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Acetylpolyamine amidohydrolase, PHOSPHATE ION, ... | | Authors: | Lombardi, P.M, Christianson, D.W. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of prokaryotic polyamine deacetylase reveals evolutionary functional relationships with eukaryotic histone deacetylases .
Biochemistry, 50, 2011
|
|
3QHR
 
 | | Structure of a pCDK2/CyclinA transition-state mimic | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CDK2 substrate peptide: PKTPKKAKKL, CHLORIDE ION, ... | | Authors: | Young, M.A, Jacobsen, D.M, Bao, Z.Q. | | Deposit date: | 2011-01-26 | | Release date: | 2011-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Briefly Bound to Activate: Transient Binding of a Second Catalytic Magnesium Activates the Structure and Dynamics of CDK2 Kinase for Catalysis.
Structure, 19, 2011
|
|
3QIJ
 
 | | Primitive-monoclinic crystal structure of the FERM domain of protein 4.1R | | Descriptor: | Protein 4.1, UNKNOWN ATOM OR ION | | Authors: | Nedyalkova, L, Zhong, N, Tong, Y, Tempel, W, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-27 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Primitive-monoclinic crystal structure of the FERM domain of protein 4.1R
to be published
|
|
3QIZ
 
 | | Crystal Structure of BoNT/A LC complexed with Hydroxamate-based Inhibitor PT-2 | | Descriptor: | (2S,4R)-2-(2-{[3-(4-fluoro-3-methylphenyl)propyl](methyl)amino}ethyl)-4-(4-fluorophenyl)-N-hydroxy-4-methoxybutanamide, Botulinum neurotoxin type A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Thompson, A.A, Han, G.W, Stevens, R.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Three Novel Hydroxamate-Based Zinc Chelating Inhibitors of the Clostridium botulinum Serotype A Neurotoxin Light Chain Metalloprotease Reveals a Compact Binding Site Resulting from 60/70 Loop Flexibility.
Biochemistry, 50, 2011
|
|
3QJF
 
 | | Crystal structure of the 2B4 TCR | | Descriptor: | 2B4 alpha chain, 2B4 beta chain | | Authors: | Ely, L.K, Newell, E.W, Davis, M.M, Garcia, K.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-27 | | Last modified: | 2011-08-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k).
J.Immunol., 186, 2011
|
|
3QC2
 
 | |
3QK0
 
 | | Crystal structure of PI3K-gamma in complex with benzothiazole 82 | | Descriptor: | N-[6-(6-chloro-5-{[(4-fluorophenyl)sulfonyl]amino}pyridin-3-yl)-1,3-benzothiazol-2-yl]acetamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery and Optimization of a Series of Benzothiazole Phosphoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QCM
 
 | |
3QD3
 
 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 1,1-Dimethylethyl {(3R,6S)-1-[2-amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-6-methyl-3-piperidinyl}carbamate | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3QLW
 
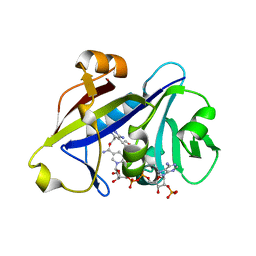 | | Candida albicans dihydrofolate reductase complexed with NADPH and 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine (UCP120B) | | Descriptor: | 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative uncharacterized protein CaJ7.0360 | | Authors: | Paulsen, J.L, Bendel, S.D, Anderson, A.C. | | Deposit date: | 2011-02-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Crystal Structures of Candida albicans Dihydrofolate Reductase Bound to Propargyl-Linked Antifolates Reveal the Flexibility of Active Site Loop Residues Critical for Ligand Potency and Selectivity.
Chem.Biol.Drug Des., 78, 2011
|
|
3QEQ
 
 | |
3QFJ
 
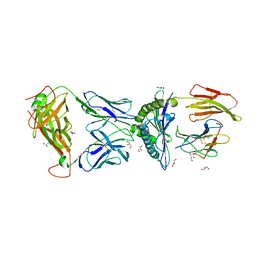 | | The complex between TCR A6 and human Class I MHC HLA-A2 with the modified TAX (Y5F) peptide | | Descriptor: | A6 alpha chain, A6 beta chain, Beta-2-microglobulin, ... | | Authors: | Borbulevych, O.Y, Baker, B.M. | | Deposit date: | 2011-01-21 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Disparate degrees of hypervariable loop flexibility control T-cell receptor cross-reactivity, specificity, and binding mechanism.
J.Mol.Biol., 414, 2011
|
|
3QQO
 
 | |
3QGA
 
 | |
3QJ0
 
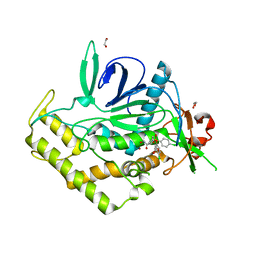 | | Crystal Structure of BoNT/A LC complexed with Hydroxamate-based Inhibitor PT-3 | | Descriptor: | (4R)-4-(4-chlorophenoxy)-1-[(4-chlorophenyl)sulfonyl]-N-hydroxy-L-prolinamide, 1,2-ETHANEDIOL, Botulinum neurotoxin type A, ... | | Authors: | Thompson, A.A, Han, G.W, Stevens, R.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural Characterization of Three Novel Hydroxamate-Based Zinc Chelating Inhibitors of the Clostridium botulinum Serotype A Neurotoxin Light Chain Metalloprotease Reveals a Compact Binding Site Resulting from 60/70 Loop Flexibility.
Biochemistry, 50, 2011
|
|
3QJH
 
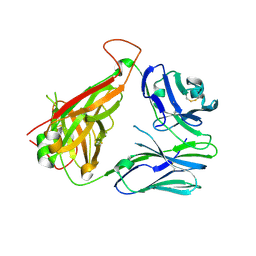 | | The crystal structure of the 5c.c7 TCR | | Descriptor: | 5c.c7 alpha chain, 5c.c7 beta chain | | Authors: | Ely, L.K, Newell, E.W, Davis, M.M, Garcia, K.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-27 | | Last modified: | 2011-08-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k).
J.Immunol., 186, 2011
|
|
3QK1
 
 | | Crystal Structure of Enterokinase-like Trypsin Variant | | Descriptor: | 1,2-ETHANEDIOL, BENZAMIDINE, CALCIUM ION, ... | | Authors: | Schoepfel, M, Tziridis, A, Arnold, U, Stubbs, M.T. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Towards a restriction proteinase: construction of a self-activating enzyme.
Chembiochem, 12, 2011
|
|
3QK9
 
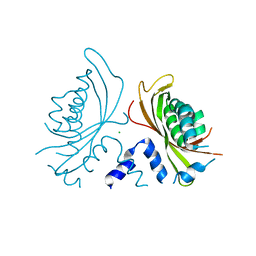 | | Yeast Tim44 C-terminal domain complexed with Cymal-3 | | Descriptor: | CHLORIDE ION, Mitochondrial import inner membrane translocase subunit TIM44 | | Authors: | Cui, W, Josyula, R, Fu, Z, Sha, B. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Membrane Binding Mechanism of Yeast Mitochondrial Peripheral Membrane Protein TIM44.
Protein Pept.Lett., 18, 2011
|
|
3Q60
 
 | |
3QXR
 
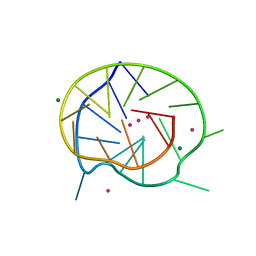 | |
3Q6K
 
 | | Salivary protein from Lutzomyia longipalpis | | Descriptor: | 43.2 kDa salivary protein, CITRIC ACID, SEROTONIN | | Authors: | Andersen, J.F, Xu, X, Chang, B.W, Collin, N, Valenzuela, J.G, Ribeiro, J.M. | | Deposit date: | 2011-01-02 | | Release date: | 2011-07-27 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure and function of a "yellow" protein from saliva of the sand fly Lutzomyia longipalpis that confers protective immunity against Leishmania major infection.
J.Biol.Chem., 286, 2011
|
|
3Q5V
 
 | |
