4AUC
 
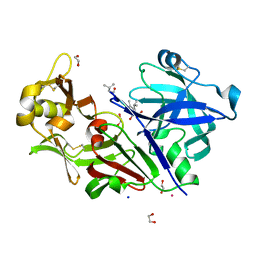 | | Bovine chymosin in complex with Pepstatin A | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHYMOSIN, ... | | Authors: | Langholm Jensen, J, Navarro Poulsen, J.C, Jacobsen, J, van den Brink, J.M, Qvist, K.B, Larsen, S. | | Deposit date: | 2012-05-16 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of Bovine Chymosin in Complex with Pepstatin A
To be Published
|
|
4GJ7
 
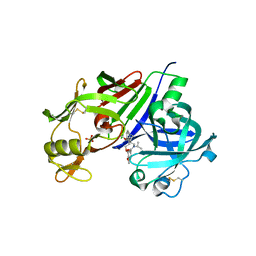 | | Crystal structure of renin in complex with NVP-BCA079 (compound 12a) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-{[(3S,4S)-4-benzylpyrrolidin-3-yl]methyl}-4-methoxy-3-(3-methoxypropoxy)-N-(propan-2-yl)benzamide, Renin, ... | | Authors: | Ostermann, N, Zink, F, Kroemer, M. | | Deposit date: | 2012-08-09 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Discovery of Novel Potent trans-3,4-Disubstituted Pyrrolidine Inhibitors of the Human Aspartic Protease Renin from in Silico Three-Dimensional (3D) Pharmacophore Searches.
J.Med.Chem., 56, 2013
|
|
4B3W
 
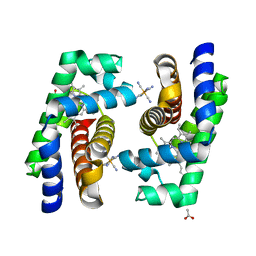 | | Crystal structure of human cytoglobin H(E7)Q mutant | | Descriptor: | ACETATE ION, CYANIDE ION, CYTOGLOBIN, ... | | Authors: | Gabba, M, Abbruzzetti, S, Spyrakis, F, Forti, F, Bruno, S, Mozzarelli, A, Luque, F.J, Viappiani, C, Cozzini, P, Nardini, M, Germani, F, Bolognesi, M, Moens, L, Dewilde, S. | | Deposit date: | 2012-07-26 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Co Rebinding Kinetics and Molecular Dynamics Simulations Highlight Dynamic Regulation of Internal Cavities in Human Cytoglobin.
Plos One, 8, 2013
|
|
4GJB
 
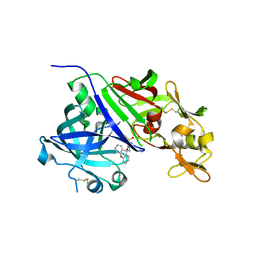 | | Crystal structure of renin in complex with NVP-BBV031 (compound 6) | | Descriptor: | (3S)-N-(9H-xanthen-9-ylmethyl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, ... | | Authors: | Ostermann, N, Zink, F, Kroemer, M. | | Deposit date: | 2012-08-09 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A novel class of oral direct Renin inhibitors: highly potent 3,5-disubstituted piperidines bearing a tricyclic p3-p1 pharmacophore.
J.Med.Chem., 56, 2013
|
|
3EXO
 
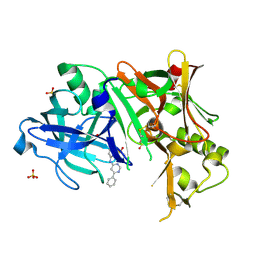 | | Crystal structure of BACE1 bound to inhibitor | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{2-methyl-5-[(6-phenylpyrimidin-4-yl)amino]phenyl}methanesulfonamide, ... | | Authors: | Allison, T.J. | | Deposit date: | 2008-10-16 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of a small molecule beta-secretase inhibitor that binds without catalytic aspartate engagement.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3EXX
 
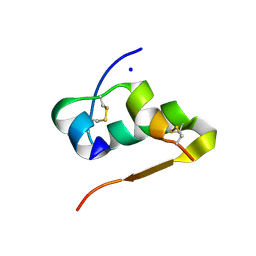 | |
4G3R
 
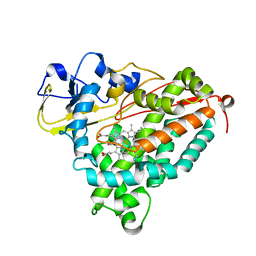 | | Crystal Structure of Nitrosyl Cytochrome P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, NITRIC OXIDE, ... | | Authors: | Madrona, Y, Tripathi, S.M, Li, H, Poulos, T.L. | | Deposit date: | 2012-07-15 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of substrate-free and nitrosyl cytochrome p450cin: implications for o(2) activation.
Biochemistry, 51, 2012
|
|
4GL5
 
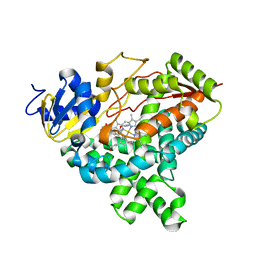 | |
4BCZ
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form. | | Descriptor: | SUPEROXIDE DISMUTASE [CU-ZN] | | Authors: | Saraboji, K, Awad, W, Danielsson, J, Lang, L, Kurnik, M, Marklund, S.L, Oliveberg, M, Logan, D.T. | | Deposit date: | 2012-10-03 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Global Structural Motions from the Strain of a Single Hydrogen Bond.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4G90
 
 | | Crystal structure of Ribonuclease A in complex with 5e | | Descriptor: | 1-{[1-(alpha-L-arabinofuranosyl)-1H-1,2,3-triazol-4-yl]methyl}-5-fluoro-2,4-dioxo-1,2,3,4-tetrahydropyrimidine, Ribonuclease pancreatic | | Authors: | Chatzileontiadou, D.S.M, Kantsadi, A.L, Leonidas, D.D. | | Deposit date: | 2012-07-23 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Triazole pyrimidine nucleosides as inhibitors of Ribonuclease A. Synthesis, biochemical, and structural evaluation.
Bioorg.Med.Chem., 20, 2012
|
|
4BEL
 
 | | BACE2 XAPERONE COMPLEX | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-SECRETASE 2, CHLORIDE ION, ... | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-03-11 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4FUJ
 
 | | Crystal Structure of the Urokinase | | Descriptor: | 6-{(E)-2-[3-(2-hydroxyethyl)phenyl]ethenyl}naphthalene-2-carboximidamide, ACETATE ION, GLYCEROL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Nienaber, V, Giranda, V. | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of the Urokinase
TO BE PUBLISHED
|
|
4AZQ
 
 | | Murine epidermal fatty acid-binding protein (FABP5) in complex with the endocannabinoid 2-arachidonoylglycerol | | Descriptor: | 2-hydroxy-1-(hydroxymethyl)ethyl icosanoate, CHLORIDE ION, FATTY ACID-BINDING PROTEIN, ... | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4GB1
 
 | | Synthesis and Evaluation of Novel 3-C-alkylated-Neu5Ac2en Derivatives as Probes of Influenza Virus Sialidase 150-loop flexibility | | Descriptor: | 5-acetamido-2,6-anhydro-3,5-dideoxy-3-[(2E)-3-phenylprop-2-en-1-yl]-D-glycero-L-altro-non-2-enonic acid, CALCIUM ION, Neuraminidase | | Authors: | Kerry, P.S, Rudrawar, S, Rameix-Welti, M.-A, Maggioni, A, Dyason, J.C, Rose, F.J, van der Werf, S, Thomson, R.J, Naffakh, N, Russell, R.J.M, von Itzstein, M. | | Deposit date: | 2012-07-26 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Synthesis and evaluation of novel 3-C-alkylated-Neu5Ac2en derivatives as probes of influenza virus sialidase 150-loop flexibility.
Org.Biomol.Chem., 10, 2012
|
|
4IGA
 
 | |
4AZY
 
 | | Design and Synthesis of BACE1 Inhibitors with In Vivo Brain Reduction of beta-Amyloid Peptides (COMPOUND 10) | | Descriptor: | (1S)-4-fluoro-1-(4-fluoro-3-pyrimidin-5-ylphenyl)-1-[2-(trifluoromethyl)pyridin-4-yl]-1H-isoindol-3-amine, ACETATE ION, BETA-SECRETASE 1, ... | | Authors: | Swahn, B.M, Kolmodin, K, Karlstrom, S, von Berg, S, Soderman, P, Holenz, J, Berg, S, Lindstrom, J, Sundstrom, M, Turek, D, Kihlstrom, J, Slivo, C, Andersson, L, Pyring, D, Ohberg, L, Kers, A, Bogar, K, Bergh, M, Olsson, L.L, Janson, J, Eketjall, S, Georgievska, B, Jeppsson, F, Falting, J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Design and synthesis of beta-site amyloid precursor protein cleaving enzyme (BACE1) inhibitors with in vivo brain reduction of beta-amyloid peptides.
J. Med. Chem., 55, 2012
|
|
4INS
 
 | | THE STRUCTURE OF 2ZN PIG INSULIN CRYSTALS AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | INSULIN (CHAIN A), INSULIN (CHAIN B), ZINC ION | | Authors: | Dodson, G.G, Dodson, E.J, Hodgkin, D.C, Isaacs, N.W, Vijayan, M. | | Deposit date: | 1989-07-10 | | Release date: | 1990-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of 2Zn pig insulin crystals at 1.5 A resolution.
Philos.Trans.R.Soc.London,Ser.B, 319, 1988
|
|
3F8A
 
 | | Crystal Structure of the R132K:R111L:L121E:R59W Mutant of Cellular Retinoic Acid-Binding Protein Type II Complexed with C15-aldehyde (a retinal analog) at 1.95 Angstrom resolution. | | Descriptor: | 1,3,3-trimethyl-2-[(1E,3E)-3-methylpenta-1,3-dien-1-yl]cyclohexene, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellular retinoic acid-binding protein 2 | | Authors: | Jia, X, Geiger, J.H. | | Deposit date: | 2008-11-12 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing Wavelength Regulation with an Engineered Rhodopsin Mimic and a C15-Retinal Analogue
Chempluschem, 77, 2012
|
|
3F7R
 
 | |
4AVD
 
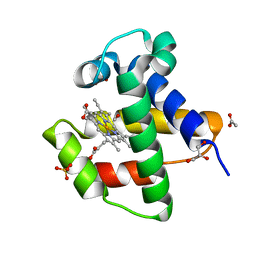 | | C.lacteus nerve Hb in complex with CO | | Descriptor: | ACETATE ION, CARBON MONOXIDE, GLYCEROL, ... | | Authors: | Germani, F, Pesce, A, Venturini, A, Moens, L, Bolognesi, M, Dewilde, S, Nardini, M. | | Deposit date: | 2012-05-25 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High Resolution Crystal Structures of the Cerebratulus Lacteus Mini-Hb in the Unligated and Carbomonoxy States.
Int.J.Mol.Sci., 13, 2012
|
|
4BFB
 
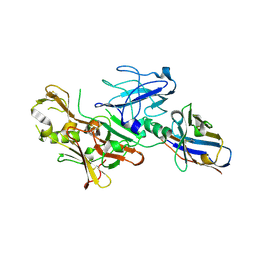 | | BACE2 XAPERONE COMPLEX | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-SECRETASE 2, XA4813 | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-03-18 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GID
 
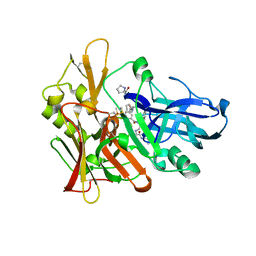 | | Structure of beta-secretase complexed with inhibitor | | Descriptor: | Beta-secretase 1, L-PROLINAMIDE, N-[(2S)-1-({(2S,3R)-3-hydroxy-1-[(2-methylpropyl)amino]-1-oxobutan-2-yl}amino)-3-phenylpropan-2-yl]-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | Authors: | Ghosh, A, Tang, J, Venkateswara, R.K, Yadav, N, Anderson, D, Gavande, N, Huang, X, Terzyan, S. | | Deposit date: | 2012-08-08 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of highly selective beta-secretase inhibitors: synthesis, biological evaluation, and protein-ligand X-ray crystal structure.
J.Med.Chem., 55, 2012
|
|
4GJ0
 
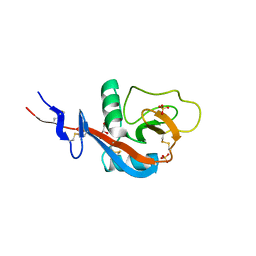 | | Crystal structure of CD23 lectin domain mutant S252A | | Descriptor: | GLYCEROL, Low affinity immunoglobulin epsilon Fc receptor, SULFATE ION | | Authors: | Yuan, D, Sutton, B.J, Dhaliwal, B. | | Deposit date: | 2012-08-09 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Ca2+-dependent Structural Changes in the B-cell Receptor CD23 Increase Its Affinity for Human Immunoglobulin E.
J.Biol.Chem., 288, 2013
|
|
4GK1
 
 | | Crystal structure of CD23 lectin domain mutant D270A | | Descriptor: | GLYCEROL, Low affinity immunoglobulin epsilon Fc receptor, SULFATE ION | | Authors: | Yuan, D, Sutton, B.J, Dhaliwal, B. | | Deposit date: | 2012-08-10 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.236 Å) | | Cite: | Ca2+-dependent Structural Changes in the B-cell Receptor CD23 Increase Its Affinity for Human Immunoglobulin E.
J.Biol.Chem., 288, 2013
|
|
4FSL
 
 | |
