6LFE
 
 | | Rat-COMT, Nitecapone,SAM and Mg bond | | Descriptor: | 3-(3,4-dihydroxy-5-nitrobenzylidene)pentane-2,4-dione, Catechol O-methyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Takebe, K, Iijima, H, Suzuki, M. | | Deposit date: | 2019-12-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Catechol O-Methyltransferase Complexed with Nitecapone.
Chem Pharm Bull (Tokyo), 68, 2020
|
|
6LFR
 
 | | Poa1p in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose 1''-phosphate phosphatase | | Authors: | Chiu, Y.C, Hsu, C.H. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Expanding the Substrate Specificity of Macro Domains toward 3''-Isomer of O-Acetyl-ADP-ribose
Acs Catalysis, 11, 2021
|
|
6LGV
 
 | | Crystal structure of a cysteine-pair mutant (P10C-S291C) of a bacterial bile acid transporter in an inward-facing state complexed with citrate | | Descriptor: | 2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRIC ACID, Transporter, ... | | Authors: | Wang, X, Lyu, Y, Ji, Y, Sun, Z, Zhou, X. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Substrate binding in the bile acid transporter ASBT Yf from Yersinia frederiksenii.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
2ITC
 
 | | Potassium Channel KcsA-Fab complex in Sodium Chloride | | Descriptor: | Antibody Fab fragment heavy chain, Antibody Fab fragment light chain, SODIUM ION, ... | | Authors: | Lockless, S.W, Zhou, M, MacKinnon, R. | | Deposit date: | 2006-10-19 | | Release date: | 2007-05-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Thermodynamic Properties of Selective Ion Binding in a K(+) Channel.
Plos Biol., 5, 2007
|
|
6LHI
 
 | | Quadruple mutant (N51I+C59R+S108N+I164L) plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with C466 (compound 42) and NADPH | | Descriptor: | 1-[3-[(4-chlorophenyl)-(phenylmethyl)amino]propoxy]-6,6-dimethyl-1,3,5-triazine-2,4-diamine, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Vitsupakorn, D. | | Deposit date: | 2019-12-09 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Flexible diaminodihydrotriazine inhibitors of Plasmodium falciparum dihydrofolate reductase: Binding strengths, modes of binding and their antimalarial activities.
Eur.J.Med.Chem., 195, 2020
|
|
6LI4
 
 | | Crystal structure of MCR-1-S | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2019-12-10 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Resensitizing carbapenem- and colistin-resistant bacteria to antibiotics using auranofin.
Nat Commun, 11, 2020
|
|
6LG0
 
 | | Crystal structure of SbCGTa in complex with UDP | | Descriptor: | SbCGTa, URIDINE-5'-DIPHOSPHATE | | Authors: | Gao, H.M, Yun, C.H. | | Deposit date: | 2019-12-04 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Dissection of the general two-step di- C -glycosylation pathway for the biosynthesis of (iso)schaftosides in higher plants.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LH1
 
 | | Crystal structure of a cysteine-pair mutant (Y113C-P190C) of a bacterial bile acid transporter trapped in an outward-facing conformation | | Descriptor: | 2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRIC ACID, Transporter, ... | | Authors: | Wang, X, Lyu, Y, Ji, Y, Sun, Z, Zhou, X. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.861 Å) | | Cite: | An engineered disulfide bridge traps and validates an outward-facing conformation in a bile acid transporter.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6LHA
 
 | | The cryo-EM structure of coxsackievirus A16 mature virion | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHQ
 
 | | The cryo-EM structure of coxsackievirus A16 mature virion in complex with Fab NA9D7 | | Descriptor: | SPHINGOSINE, VP1 protein, VP2 protein, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LIM
 
 | | BRD4-BD1 bound with compound 40 | | Descriptor: | (3~{R})-6-[(4-isoquinolin-4-ylpyrimidin-2-yl)amino]-1,3-dimethyl-4-propan-2-yl-3~{H}-quinoxalin-2-one, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D, Li, Y. | | Deposit date: | 2019-12-12 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75909507 Å) | | Cite: | BRD4-BD1 bound with compound 40
To Be Published
|
|
6LIR
 
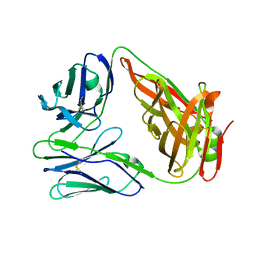 | | crystal structure of chicken TCR for 2.0 | | Descriptor: | TCR alpha chain, TCR beta chain | | Authors: | Zhang, L, Xia, C. | | Deposit date: | 2019-12-12 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural and Biophysical Insights into the TCR alpha beta Complex in Chickens.
Iscience, 23, 2020
|
|
6LI2
 
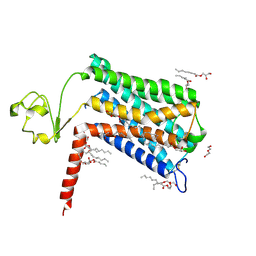 | | Crystal structure of GPR52 ligand free form with rubredoxin fusion | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera of G-protein coupled receptor 52 and Rubredoxin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
6LIH
 
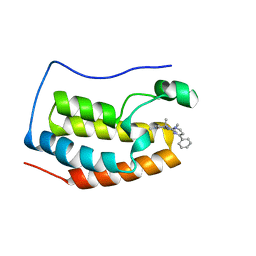 | | BRD4 BD1 bound with compound 10 | | Descriptor: | (3~{R})-1,3-dimethyl-6-[(4-phenylpyrimidin-2-yl)amino]-4-propan-2-yl-3~{H}-quinoxalin-2-one, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D. | | Deposit date: | 2019-12-11 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62030542 Å) | | Cite: | BRD4 BD1 bound with compound 10
To Be Published
|
|
6LIL
 
 | |
6LJ6
 
 | |
6LJK
 
 | |
6LKA
 
 | | Crystal Structure of EV71-3C protease with a Novel Macrocyclic Compounds | | Descriptor: | 3C proteinase, ~{N}-[(2~{S})-1-[[(2~{S},3~{S},6~{S},7~{Z},12~{E})-4,9-bis(oxidanylidene)-6-[[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]methyl]-2-phenyl-1,10-dioxa-5-azacyclopentadeca-7,12-dien-3-yl]amino]-3-methyl-1-oxidanylidene-butan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Li, P, Wu, S.Q, Xiao, T.Y.C, Li, Y.L, Su, Z.M, Hao, F, Hu, G.P, Hu, J, Lin, F.S, Chen, X.S, Gu, Z.X, He, H.Y, Li, J, Chen, S.H. | | Deposit date: | 2019-12-18 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Design, synthesis, and evaluation of a novel macrocyclic anti-EV71 agent.
Bioorg.Med.Chem., 28, 2020
|
|
6L9H
 
 | | The Human Telomeric Nucleosome Displays Distinct Structural and Dynamic Properties | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-K, Histone H3.1, ... | | Authors: | Soman, A, Liew, C.W, Teo, H.L, Berezhnoy, N, Korolev, N, Rhodes, D, Nordenskiold, L. | | Deposit date: | 2019-11-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The human telomeric nucleosome displays distinct structural and dynamic properties.
Nucleic Acids Res., 48, 2020
|
|
6LIY
 
 | | SeMet CRL Protein of Arabidopsis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Chromophore lyase CRL, chloroplastic | | Authors: | Wang, F.F, Guan, K.L, Sun, P.K, Xing, W.M. | | Deposit date: | 2019-12-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | The Arabidopsis CRUMPLED LEAF protein, a homolog of the cyanobacterial bilin lyase, retains the bilin-binding pocket for a yet unknown function.
Plant J., 104, 2020
|
|
2IDR
 
 | | Crystal structure of translation initiation factor EIF4E from wheat | | Descriptor: | Eukaryotic translation initiation factor 4E-1 | | Authors: | Monzingo, A.F, Sadow, J, Dhaliwal, S, Lyon, A, Hoffman, D.W, Robertus, J.D, Browning, K.S. | | Deposit date: | 2006-09-15 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond.
Plant Physiol., 143, 2007
|
|
6LJ4
 
 | | Crystal structure of NDM-1 in complex with D-captopril derivative wss04146 | | Descriptor: | (3S)-1-[(2S)-2-methyl-3-sulfanyl-propanoyl]piperidine-3-carboxylic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Zhang, H, Ma, G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure-guided optimization of D-captopril for discovery of potent NDM-1 inhibitors.
Bioorg.Med.Chem., 29, 2020
|
|
6LJH
 
 | | Crystal structure of Alcohol dehydrogenase 1 from Artemisia annua in complex with NAD+ | | Descriptor: | Alcohol dehydrogenase 1, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Feng, X, Fan, S, Li, M, Zou, S, Lv, G, Wu, G, Jin, Y, Yang, Z. | | Deposit date: | 2019-12-16 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Alcohol dehydrogenase 1 from Artemisia annua in complex with NAD+
To Be Published
|
|
6LJV
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 2-[[3-chloranyl-2-(2,3-dihydro-1-benzofuran-5-yl)phenyl]amino]benzoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
6LA9
 
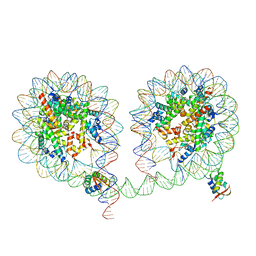 | | 349 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 (high cryoprotectant) | | Descriptor: | CALCIUM ION, DNA (349-MER), Histone H1.0, ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
