2ME9
 
 | |
8EX8
 
 | |
8F8Q
 
 | | Cryo-EM structure of the CapZ-capped barbed end of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Carman, P.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2022-11-22 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structures of the free and capped ends of the actin filament.
Science, 380, 2023
|
|
4AXG
 
 | | Structure of eIF4E-Cup complex | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, PROTEIN CUP | | Authors: | Kinkelin, K, Veith, K, Gruenwald, M, Bono, F. | | Deposit date: | 2012-06-12 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Minimal Eif4E-Cup Complex Revelas a General Mechanism of Eif4E Regulation in Translational Repression
RNA, 18, 2012
|
|
5U6H
 
 | | Solution structure of the zinc fingers 1 and 2 of MBNL1 | | Descriptor: | Muscleblind-like protein 1, ZINC ION | | Authors: | Phukan, P.D, Park, S, Martinez-Yamout, M.M, Zeeb, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-12-08 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Interaction of the Tandem Zinc Finger Domains of Human Muscleblind with Cognate RNA from Human Cardiac Troponin T.
Biochemistry, 56, 2017
|
|
8ESM
 
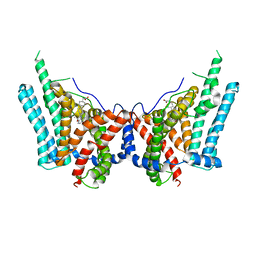 | | Human triacylglycerol synthesizing enzyme DGAT1 in complex with T863 inhibitor | | Descriptor: | Diacylglycerol O-acyltransferase 1, {(1r,4r)-4-[4-(4-amino-7,7-dimethyl-7H-pyrimido[4,5-b][1,4]oxazin-6-yl)phenyl]cyclohexyl}acetic acid | | Authors: | Sui, X, Kun, W, Walther, T, Farese, R, Liao, M. | | Deposit date: | 2022-10-14 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of action for small-molecule inhibitors of triacylglycerol synthesis.
Nat Commun, 14, 2023
|
|
5UFJ
 
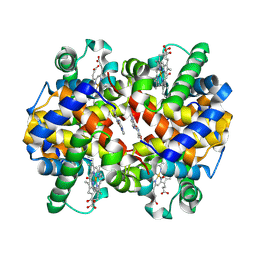 | | Crystal Structure of Carbonmonoxy Hemoglobin S (Liganded Sickle Cell Hemoglobin) Complexed with GBT Compound 6 | | Descriptor: | 5-[(imidazo[1,2-a]pyridin-8-yl)methoxy]-2-methoxypyridine-4-carbaldehyde, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Partridge, J.R, Choy, R.M, Li, Z, Metcalf, B. | | Deposit date: | 2017-01-04 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of GBT440, an Orally Bioavailable R-State Stabilizer of Sickle Cell Hemoglobin.
ACS Med Chem Lett, 8, 2017
|
|
8ETM
 
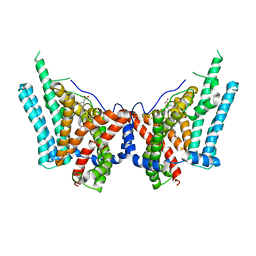 | | Human triacylglycerol synthesizing enzyme DGAT1 in complex with DGAT1IN1 inhibitor | | Descriptor: | Diacylglycerol O-acyltransferase 1, [(1S,4r)-4-{4-[(4S)-2-({[4-(trifluoromethoxy)phenyl]methyl}carbamoyl)imidazo[1,2-a]pyridin-6-yl]phenyl}cyclohexyl]acetic acid | | Authors: | Sui, X, Kun, W, Walther, T, Farese, R, Liao, M. | | Deposit date: | 2022-10-17 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of action for small-molecule inhibitors of triacylglycerol synthesis.
Nat Commun, 14, 2023
|
|
8F8R
 
 | | Cryo-EM structure of the free barbed end of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Carman, P.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2022-11-22 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of the free and capped ends of the actin filament.
Science, 380, 2023
|
|
8F8P
 
 | | Cryo-EM structure of F-actin in the ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Carman, P.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2022-11-22 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Structures of the free and capped ends of the actin filament.
Science, 380, 2023
|
|
5TUX
 
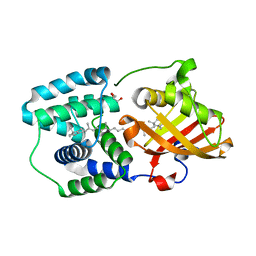 | | crystal structure and light induced structural changes in orange carotenoid protein bound with echinenone | | Descriptor: | GLYCEROL, Orange carotenoid-binding protein, beta,beta-caroten-4-one | | Authors: | Yang, X, Bandara, S, Ren, Z. | | Deposit date: | 2016-11-07 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Photoactivation mechanism of a carotenoid-based photoreceptor.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8F8S
 
 | | Cryo-EM structure of the free pointed end of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Carman, P.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2022-11-22 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structures of the free and capped ends of the actin filament.
Science, 380, 2023
|
|
6UZ3
 
 | | Cardiac sodium channel | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jiang, D, Shi, H, Tonggu, L, Lenaeus, M.J, Zheng, N, Catterall, W.A. | | Deposit date: | 2019-11-14 | | Release date: | 2020-01-01 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Cardiac Sodium Channel.
Cell, 180, 2020
|
|
8F8T
 
 | | Cryo-EM structure of the Tropomodulin-capped pointed end of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Carman, P.J, Barrie, K.R, Dominguez, R. | | Deposit date: | 2022-11-22 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structures of the free and capped ends of the actin filament.
Science, 380, 2023
|
|
2M9E
 
 | | NMR solution structure of Pin1 WW domain mutant 5-1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Enck, S, Chen, W, Price, J.L, Powers, E.T, Wong, C, Dyson, H.J, Kelly, J.W. | | Deposit date: | 2013-06-07 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
J.Am.Chem.Soc., 135, 2013
|
|
6V15
 
 | | immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen beta 72,74cit69-81, G08 TCR alpha chain, ... | | Authors: | Lim, J.J, Rossjohn, J. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The shared susceptibility epitope of HLA-DR4 binds citrullinated self-antigens and the TCR.
Sci Immunol, 6, 2021
|
|
4B1V
 
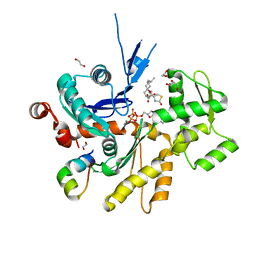 | | Structure of the Phactr1 RPEL-N domain bound to G-actin | | Descriptor: | 1,2-ETHANEDIOL, ACTIN, ALPHA SKELETAL MUSCLE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
2M03
 
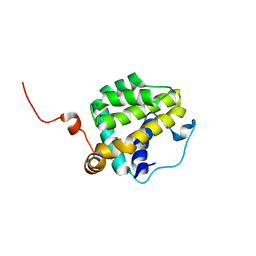 | |
4B3P
 
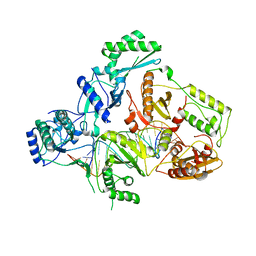 | | Structures of HIV-1 RT and RNA-DNA Complex Reveal a Unique RT Conformation and Substrate Interface | | Descriptor: | DNA, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H, ... | | Authors: | Lapkouski, M, Tian, L, Miller, J.T, Le Grice, S.F.J, Yang, W. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.839 Å) | | Cite: | Complexes of HIV-1 RT, Nnrti and RNA/DNA Hybrid Reveal a Structure Compatible with RNA Degradation
Nat.Struct.Mol.Biol., 20, 2013
|
|
2MDJ
 
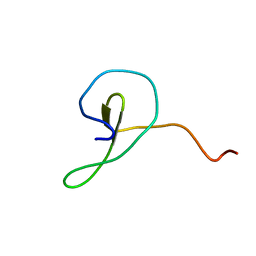 | |
5TUS
 
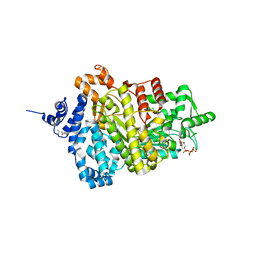 | | Potent competitive inhibition of human ribonucleotide reductase by a novel non-nucleoside small molecule | | Descriptor: | 2-hydroxy-N'-[(Z)-(2-hydroxynaphthalen-1-yl)methylidene]benzohydrazide, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Mohammed, F.A, Alam, I, Dealwis, C.G. | | Deposit date: | 2016-11-07 | | Release date: | 2017-08-02 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Potent competitive inhibition of human ribonucleotide reductase by a nonnucleoside small molecule.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TWV
 
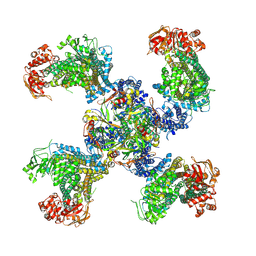 | | Cryo-EM structure of the pancreatic ATP-sensitive K+ channel SUR1/Kir6.2 in the presence of ATP and glibenclamide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ATP-sensitive inward rectifier potassium channel 11 | | Authors: | Martin, G.M, Yoshioka, C, Chen, J.Z, Shyng, S.L. | | Deposit date: | 2016-11-14 | | Release date: | 2017-01-25 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Cryo-EM structure of the ATP-sensitive potassium channel illuminates mechanisms of assembly and gating.
Elife, 6, 2017
|
|
6V8Q
 
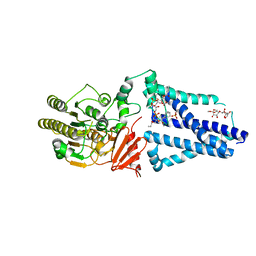 | | Structure of an inner membrane protein required for PhoPQ regulated increases in outer membrane cardiolipin | | Descriptor: | (9Z,21R,24R,30R,33R,44Z)-24,27,30-trihydroxy-18,24,30,36-tetraoxo-19,23,25,29,31,35-hexaoxa-24lambda~5~,30lambda~5~-dip hosphatripentaconta-9,44-diene-21,33-diyl (9Z,9'Z)di-octadec-9-enoate, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Fan, J, Miller, S. | | Deposit date: | 2019-12-11 | | Release date: | 2020-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.699585 Å) | | Cite: | Structure of an Inner Membrane Protein Required for PhoPQ-Regulated Increases in Outer Membrane Cardiolipin.
Mbio, 11, 2020
|
|
6UPI
 
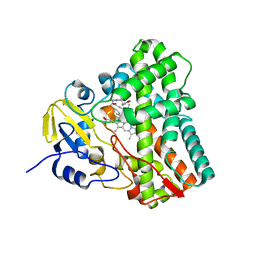 | | Crystal structure of Mycobacterium tuberculosis CYP121 bound with a hydroxylated intermediate of cYF-4-OMe | | Descriptor: | (3S,6S)-3-{[4-(hydroxymethoxy)phenyl]methyl}-6-[(4-hydroxyphenyl)methyl]piperazine-2,5-dione, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nguyen, R.C.D, Yang, Y, Liu, A. | | Deposit date: | 2019-10-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | Substrate-Assisted Hydroxylation and O-Demethylation in the Peroxidase-like Cytochrome P450 Enzyme CYP121
Acs Catalysis, 10, 2020
|
|
5U0B
 
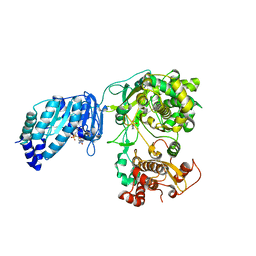 | | Structure of full-length Zika virus NS5 | | Descriptor: | Genome polyprotein, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Zhao, B, Du, F. | | Deposit date: | 2016-11-23 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and function of the Zika virus full-length NS5 protein.
Nat Commun, 8, 2017
|
|
