6T7Y
 
 | | Structure of PCNA bound to cPIP motif of DP2 from P. abyssi | | Descriptor: | DNA polymerase sliding clamp, cPIP motif from the DP2 large subunit of PolD | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
8SW0
 
 | | Puromycin sensitive aminopeptidase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Puromycin-sensitive aminopeptidase, ... | | Authors: | Rodgers, D.W, Sampath, S. | | Deposit date: | 2023-05-17 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure of puromycin-sensitive aminopeptidase and polyglutamine binding.
Plos One, 18, 2023
|
|
6NIW
 
 | | Crystal structure of P[6] rotavirus | | Descriptor: | DI(HYDROXYETHYL)ETHER, Protease-sensitive outer capsid protein | | Authors: | Xu, S, Liu, Y, Lakamp, L, Ahmed, L, Jiang, X, Kennedy, M.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular basis of P[II] major human rotavirus VP8* domain recognition of histo-blood group antigens.
Plos Pathog., 16, 2020
|
|
6O1E
 
 | | The crystal structure of human MORC3 ATPase-CW in complex with AMPPNP | | Descriptor: | MAGNESIUM ION, MORC family CW-type zinc finger protein 3, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Klein, B.J, Zhang, Y, Kutateladze, T.G. | | Deposit date: | 2019-02-19 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Mechanism for autoinhibition and activation of the MORC3 ATPase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6JW4
 
 | | Degenerate RVD RG forms a distinct loop conformation | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5HC)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | Authors: | Liu, L, Yi, C. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
6S83
 
 | | Crystal structure of methionine adenosyltransferase from Pyrococcus furiosus in complex with AMPPCP, SAM, and PCP | | Descriptor: | MAGNESIUM ION, METHYLENEDIPHOSPHONIC ACID, PHOSPHATE ION, ... | | Authors: | Degano, M, Minici, C, Porcelli, M. | | Deposit date: | 2019-07-08 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.336 Å) | | Cite: | Structures of catalytic cycle intermediates of the Pyrococcus furiosus methionine adenosyltransferase demonstrate negative cooperativity in the archaeal orthologues.
J.Struct.Biol., 210, 2020
|
|
6JW5
 
 | | RVD Q* recognizes 5hmC through water-mediated H bonds | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5HC)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | Authors: | Liu, L, Yi, C. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
6T8N
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K3007 | | Descriptor: | 1,2-ETHANEDIOL, Activin receptor type I, DIMETHYL SULFOXIDE, ... | | Authors: | Adamson, R.J, Williams, E.P, Bonomo, S, Rankin, S, Bacos, D, Rae, A, Cramp, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-10-24 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K3007
To Be Published
|
|
6JW0
 
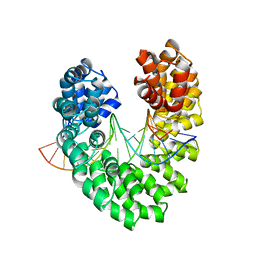 | | Universal RVD R* accommodates cytosine via water-mediated interactions | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*CP*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | Authors: | Liu, L, Yi, C. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
6NV2
 
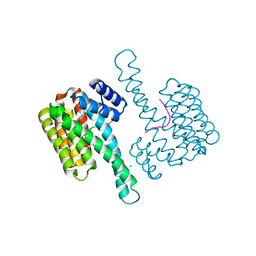 | | 14-3-3 sigma with RelA/p65 binding site pS45 in complex with DP005 | | Descriptor: | (4R,5R,6R,6aS,9S,9aE,10aR)-5-hydroxy-9-(methoxymethyl)-6,10a-dimethyl-3-(propan-2-yl)-1,2,4,5,6,6a,7,8,9,10a-decahydrodicyclopenta[a,d][8]annulen-4-yl alpha-D-glucopyranoside, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2019-02-04 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Selectivity via Cooperativity: Preferential Stabilization of the p65/14-3-3 Interaction with Semisynthetic Natural Products.
J.Am.Chem.Soc., 142, 2020
|
|
6JW3
 
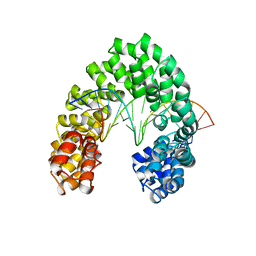 | | Degenerate RVD RG forms a distinct loop conformation | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5CM)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | Authors: | Liu, L, Yi, C. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
6KG9
 
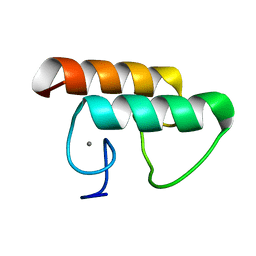 | | Solution structure of CaDoc0917 from Clostridium acetobutylicum | | Descriptor: | And cellulose-binding endoglucanase family 9 CelL ortholog dockerin domain, CALCIUM ION | | Authors: | Feng, Y, Yao, X. | | Deposit date: | 2019-07-11 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules.
Sci Adv, 6, 2020
|
|
6TLF
 
 | | human 14-3-3 sigma isoform in complex with IMP | | Descriptor: | 14-3-3 protein sigma, INOSINIC ACID, SULFATE ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S. | | Deposit date: | 2019-12-02 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of Phosphate-Containing Compounds as New Inhibitors of 14-3-3/c-Abl Protein-Protein Interaction.
Acs Chem.Biol., 15, 2020
|
|
8Q7I
 
 | |
8Q7S
 
 | | Crystal structure of the SARS-CoV-2 RBD (Wuhan) with neutralizing VHHs Ma6F06 and Re21H01 | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, GLYCEROL, ... | | Authors: | Guttler, T, Aksu, M, Gorlich, D. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
6JW1
 
 | | Universal RVD R* accommodates 5mC via water-mediated interactions | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5CM)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | Authors: | Liu, L, Yi, C. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
8I02
 
 | | Cryo-EM structure of the SIN3S complex from S. pombe | | Descriptor: | Chromatin modification-related protein eaf3, Cph1, Histone deacetylase clr6, ... | | Authors: | Wang, C, Guo, Z, Zhan, X. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Two assembly modes for SIN3 histone deacetylase complexes.
Cell Discov, 9, 2023
|
|
6TM7
 
 | | Human 14-3-3 sigma isoform in complex with PLP | | Descriptor: | 14-3-3 protein sigma, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S. | | Deposit date: | 2019-12-03 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of Phosphate-Containing Compounds as New Inhibitors of 14-3-3/c-Abl Protein-Protein Interaction.
Acs Chem.Biol., 15, 2020
|
|
8TIM
 
 | |
8TXS
 
 | |
6TWZ
 
 | | 14-3-3 sigma complexed with a phosphorylated 16E6 peptide | | Descriptor: | 14-3-3 protein sigma, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D(-)-TARTARIC ACID, ... | | Authors: | Gogl, G, Cousido-Siah, A, Sluchanko, N.N, Trave, G. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual Specificity PDZ- and 14-3-3-Binding Motifs: A Structural and Interactomics Study.
Structure, 28, 2020
|
|
6JW2
 
 | | Universal RVD R* accommodates 5hmC via water-mediated interactions | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5HC)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | Authors: | Liu, L, Yi, C. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
6JVZ
 
 | | RVD HA specifically contacts 5mC through van der Waals interactions | | Descriptor: | DNA (5'-D(*AP*GP*AP*GP*AP*CP*GP*CP*GP*AP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*(5CM)P*GP*CP*GP*TP*CP*TP*CP*T)-3'), TAL effector | | Authors: | Liu, L, Yi, C. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural Insights into the Specific Recognition of 5-methylcytosine and 5-hydroxymethylcytosine by TAL Effectors.
J.Mol.Biol., 432, 2020
|
|
5M32
 
 | | Human 26S proteasome in complex with Oprozomib | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Haselbach, D, Schrader, J, Lambrecht, F, Henneberg, F, Chari, A, Stark, H. | | Deposit date: | 2016-10-14 | | Release date: | 2017-07-05 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Long-range allosteric regulation of the human 26S proteasome by 20S proteasome-targeting cancer drugs.
Nat Commun, 8, 2017
|
|
4BGV
 
 | |
