5U0F
 
 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-[(2,4-dimethoxyphenyl)methyl]-2-sulfanylidene-1,3-thiazolidin-4-one, Carbonic anhydrase 2, ZINC ION | | Authors: | Peat, T.S, Poulsen, S.A, Ren, B, Dolezal, O, Woods, L.A, Mujumdar, P, Chrysanthopoulos, P.K. | | Deposit date: | 2016-11-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
8ONL
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A | | Descriptor: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
6MIW
 
 | | WWE domain of human HUWE1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, E3 ubiquitin-protein ligase HUWE1, UNKNOWN ATOM OR ION | | Authors: | Halabelian, L, Loppnau, P, Tempel, W, Wong, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-20 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | WWE domain of human HUWE1
To Be Published
|
|
7PLD
 
 | |
5U1V
 
 | |
8OTM
 
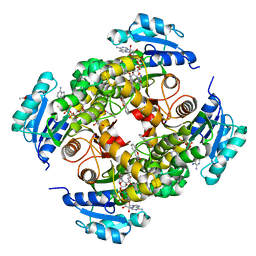 | | structure of InhA from mycobacterium tuberculosis in complex with N-((1-(3-hydroxy-4-phenoxybenzyl)-1H-1,2,3-triazol-4-yl)methyl)-2-oxo-2H-chromene-3-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 2-oxidanylidene-~{N}-[[1-[(3-oxidanyl-4-phenoxy-phenyl)methyl]-1,2,3-triazol-4-yl]methyl]chromene-3-carboxamide, ACETATE ION, ... | | Authors: | Chebaiki, M, Maveyraud, L, Tamhaev, R, Lherbet, C, Mourey, L. | | Deposit date: | 2023-04-21 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of new diaryl ether inhibitors against Mycobacterium tuberculosis targeting the minor portal of InhA.
Eur.J.Med.Chem., 259, 2023
|
|
5C98
 
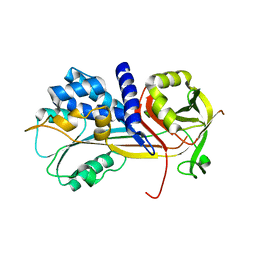 | | 1.45A resolution structure of SRPN18 from Anopheles gambiae | | Descriptor: | AGAP007691-PB | | Authors: | Lovell, S, Battaile, K.P, Gulley, M, Zhang, X, Meekins, D.A, Gao, F.P, Michel, K. | | Deposit date: | 2015-06-26 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 angstrom resolution structure of SRPN18 from the malaria vector Anopheles gambiae.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
7Z5H
 
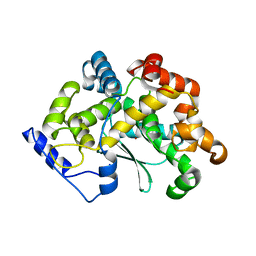 | | human Zn MATCAP | | Descriptor: | Uncharacterized protein KIAA0895-like, ZINC ION | | Authors: | Bak, J, Adamopoulos, A, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Posttranslational modification of microtubules by the MATCAP detyrosinase.
Science, 376, 2022
|
|
7Z5G
 
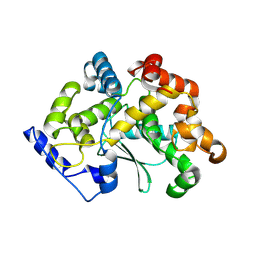 | | human apo MATCAP | | Descriptor: | Uncharacterized protein KIAA0895-like | | Authors: | Bak, J, Adamoupolos, A, Heidebrecht, T, Perrakis, A. | | Deposit date: | 2022-03-09 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Posttranslational modification of microtubules by the MATCAP detyrosinase.
Science, 376, 2022
|
|
8ONJ
 
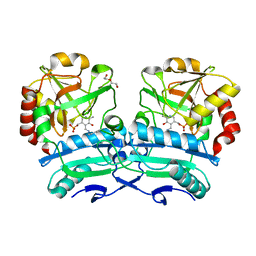 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant R88L | | Descriptor: | Aminotransferase class IV, DI(HYDROXYETHYL)ETHER, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2023-04-03 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
6MH3
 
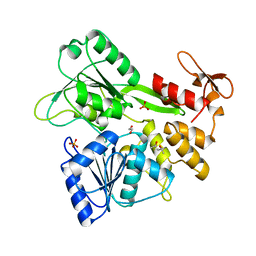 | | The crystal structure of Zika virus NS3 helicase domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, PHOSPHATE ION, ... | | Authors: | Oliva, G, Mesquita, N, Godoy, A. | | Deposit date: | 2018-09-17 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The crystal structure of Zika virus NS3 helicase domain
To Be Published
|
|
8RUJ
 
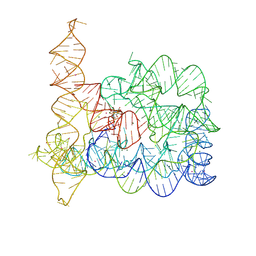 | | Structure of Oceanobacillus iheyensis group II intron in the presence of K+, Mg2+, 5'-exon, and ARN25850 after 1h soaking | | Descriptor: | 2-[2,6-bis(bromanyl)-3,4,5-tris(oxidanyl)phenyl]carbonyl-~{N}-(2-pyrrolidin-1-ylethyl)-1-benzofuran-5-carboxamide, Domains 1-5, MAGNESIUM ION, ... | | Authors: | Silvestri, I, Marcia, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Targeting the conserved active site of splicing machines with specific and selective small molecule modulators.
Nat Commun, 15, 2024
|
|
7PQ6
 
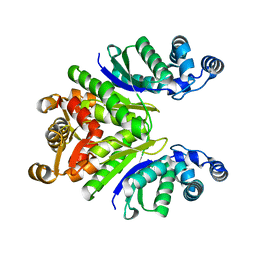 | | Crystal Structure of the Ring Nuclease 0811 mutant-S12A from Sulfolobus islandicus (Sis0811) | | Descriptor: | CRISPR-associated protein, APE2256 family | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
5CKU
 
 | |
7PLC
 
 | | Caulobacter crescentus xylonolactonase with D-xylose, P21 space group | | Descriptor: | FE (II) ION, SULFATE ION, Smp-30/Cgr1 family protein, ... | | Authors: | Paakkonen, J, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2021-08-30 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Three-dimensional structure of xylonolactonase from Caulobacter crescentus: A mononuclear iron enzyme of the 6-bladed beta-propeller hydrolase family.
Protein Sci., 31, 2022
|
|
7NL5
 
 | | Structure of the catalytic domain of the Bacillus circulans alpha-1,6 Mannanase in complex with an alpha-1,6-alpha-manno-cyclophellitol trisaccharide inhibitor | | Descriptor: | (1R,2R,3R,4S,5R)-4-(hydroxymethyl)cyclohexane-1,2,3,5-tetrol, (1R,6S)-5beta-(Hydroxymethyl)-7-oxabicyclo[4.1.0]heptane-2beta,3beta,4alpha-triol, Alpha-1,6-mannanase, ... | | Authors: | Schroeder, S, Offen, W.A, Males, A, Jin, Y, De Boer, C, Enotarpi, J, Marino, L, van der Marel, G.A, Florea, B.I, Codee, J.D.C, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Development of Non-Hydrolysable Oligosaccharide Activity-Based Inactivators for Endoglycanases: A Case Study on alpha-1,6 Mannanases.
Chemistry, 27, 2021
|
|
8OEM
 
 | | Crystal structure of FutA bound to Fe(II) | | Descriptor: | FE (II) ION, Putative iron ABC transporter, substrate binding protein | | Authors: | Bolton, R, Tews, I. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A redox switch allows binding of Fe(II) and Fe(III) ions in the cyanobacterial iron-binding protein FutA from Prochlorococcus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7VEK
 
 | | Crystal structure of Phytolacca americana UGT3 with capsaicin and UDP-2fluoroglucose | | Descriptor: | (6E)-N-(4-hydroxy-3-methoxybenzyl)-8-methylnon-6-enamide, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Glycosyltransferase, ... | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for substrate recognition in the Phytolacca americana glycosyltransferase PaGT3.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8ORA
 
 | | Human holo aromatic L-amino acid decarboxylase (AADC) external aldimine with L-Dopa methylester | | Descriptor: | Dopa decarboxylase (Aromatic L-amino acid decarboxylase), PYRIDOXAL-5'-PHOSPHATE, TETRAETHYLENE GLYCOL, ... | | Authors: | Bisello, G, Perduca, M, Bertoldi, M. | | Deposit date: | 2023-04-13 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human aromatic amino acid decarboxylase is an asymmetric and flexible enzyme: Implication in aromatic amino acid decarboxylase deficiency.
Protein Sci., 32, 2023
|
|
5U77
 
 | | Crystal structure of ORP8 PH domain | | Descriptor: | FORMIC ACID, N-(2-hydroxyethyl)-N,N-dimethyl-3-sulfopropan-1-aminium, Oxysterol-binding protein-related protein 8 | | Authors: | Ghai, R, Yang, H. | | Deposit date: | 2016-12-11 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | ORP5 and ORP8 bind phosphatidylinositol-4, 5-biphosphate (PtdIns(4,5)P 2) and regulate its level at the plasma membrane.
Nat Commun, 8, 2017
|
|
5GUV
 
 | | The crystal structure of mouse DNMT1 (731-1602) mutant - R1279D | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, ZINC ION | | Authors: | Ye, F, Chen, S.J. | | Deposit date: | 2016-08-31 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.078 Å) | | Cite: | Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1.
ACS Chem. Biol., 13, 2018
|
|
5CAW
 
 | |
7PQA
 
 | | Crystal Structure of the Ring Nuclease 0811 mutant-S12G/K169G from Sulfolobus islandicus (Sis0811) | | Descriptor: | CRISPR-associated protein, APE2256 family | | Authors: | Molina, R, Jensen, A.L.G, Marchena-Hurtado, J, Lopez-Mendez, B, Stella, S, Montoya, G. | | Deposit date: | 2021-09-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural basis of cyclic oligoadenylate degradation by ancillary Type III CRISPR-Cas ring nucleases.
Nucleic Acids Res., 49, 2021
|
|
8OWO
 
 | | SMYD3 in complex with fragment FL01507 | | Descriptor: | 3-oxidanylbenzenecarbonitrile, GLYCEROL, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Lund, B.A, Cederfelt, D, Dobritzsch, D. | | Deposit date: | 2023-04-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of fragments targeting SMYD3 using highly sensitive kinetic and multiplexed biosensor-based screening.
Rsc Med Chem, 15, 2024
|
|
7ZCL
 
 | | Unspecific peroxygenase from Collariella virescens | | Descriptor: | Collariella virescens UPO, HEME C, MAGNESIUM ION | | Authors: | Santillana, E, Romero, A. | | Deposit date: | 2022-03-28 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Characterization of Two Short Unspecific Peroxygenases: Two Different Dimeric Arrangements.
Antioxidants, 11, 2022
|
|
