3H9V
 
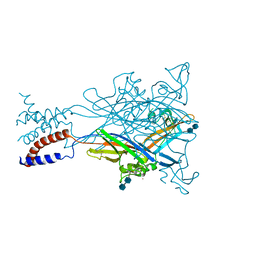 | | Crystal structure of the ATP-gated P2X4 ion channel in the closed, apo state at 3.1 Angstroms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GADOLINIUM ATOM, P2X purinoceptor | | Authors: | Kawate, T, Michel, J.C, Gouaux, E. | | Deposit date: | 2009-04-30 | | Release date: | 2009-07-28 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the ATP-gated P2X(4) ion channel in the closed state.
Nature, 460, 2009
|
|
3I5D
 
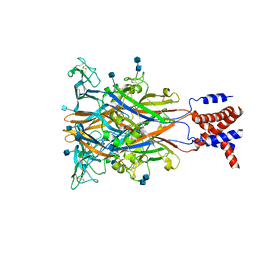 | | Crystal structure of the ATP-gated P2X4 ion channel in the closed, apo state at 3.5 Angstroms (R3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P2X purinoceptor | | Authors: | Kawate, T, Michel, J.C, Gouaux, E. | | Deposit date: | 2009-07-05 | | Release date: | 2009-08-04 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Crystal structure of the ATP-gated P2X(4) ion channel in the closed state.
Nature, 460, 2009
|
|
6VD7
 
 | |
2A65
 
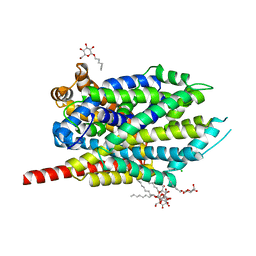 | | Crystal structure of LEUTAA, a bacterial homolog of Na+/Cl--dependent neurotransmitter transporters | | Descriptor: | CHLORIDE ION, LEUCINE, Na(+):neurotransmitter symporter (Snf family), ... | | Authors: | Yamashita, A, Singh, S.K, Kawate, T, Jin, Y, Gouaux, E. | | Deposit date: | 2005-07-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a bacterial homologue of Na(+)/Cl(-)-dependent neurotransmitter transporters.
Nature, 437, 2005
|
|
5U2H
 
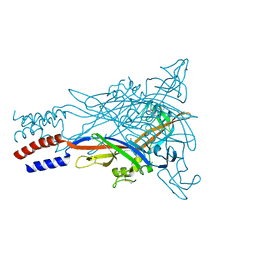 | |
5U1U
 
 | |
5U1Y
 
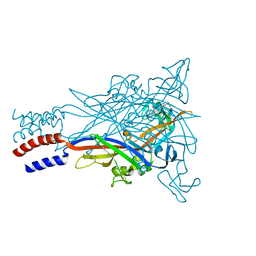 | |
5U1X
 
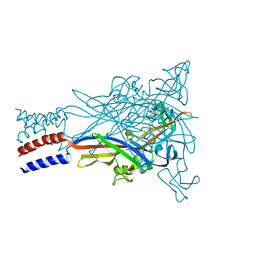 | |
5U1L
 
 | |
5U1V
 
 | |
5U1W
 
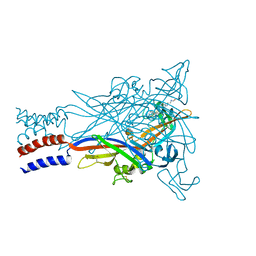 | | Crystal structure of the ATP-gated P2X7 ion channel bound to allosteric antagonist AZ10606120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-[2-({2-[(2-hydroxyethyl)amino]ethyl}amino)quinolin-5-yl]-2-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]decan-1-yl]acetamide, ... | | Authors: | Karasawa, A, Kawate, T. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Structural basis for subtype-specific inhibition of the P2X7 receptor.
Elife, 5, 2016
|
|
