1P02
 
 | |
4X53
 
 | | Structure of the class D Beta-Lactamase OXA-160 V130D in Acyl-Enzyme Complex with Aztreonam | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, BICARBONATE ION, Class D beta-lactamase OXA-160 | | Authors: | Clasman, J.R, June, C.M, Powers, R.A, Leonard, D.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Activity against Aztreonam and Extended Spectrum Cephalosporins for Two Carbapenem-Hydrolyzing Class D beta-Lactamases from Acinetobacter baumannii.
Biochemistry, 54, 2015
|
|
4X56
 
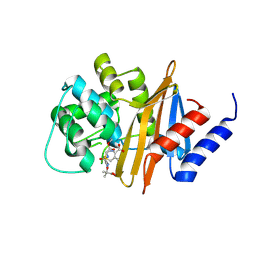 | | Structure of the class D Beta-Lactamase OXA-160 V130D in Acyl-Enzyme Complex with Ceftazidime | | Descriptor: | ACYLATED CEFTAZIDIME, Class D beta-lactamase OXA-160 | | Authors: | Clasman, J.R, June, C.M, Powers, R.A, Leonard, D.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Basis of Activity against Aztreonam and Extended Spectrum Cephalosporins for Two Carbapenem-Hydrolyzing Class D beta-Lactamases from Acinetobacter baumannii.
Biochemistry, 54, 2015
|
|
4YU5
 
 | | Crystal structure of selenomethionine variant of Bacillus anthracis immune inhibitor A2 peptidase zymogen | | Descriptor: | 3-(1-methylpiperidinium-1-yl)propane-1-sulfonate, CALCIUM ION, GLYCEROL, ... | | Authors: | Arolas, J.L, Goulas, T, Gomis-Ruth, F.X. | | Deposit date: | 2015-03-18 | | Release date: | 2015-10-28 | | Last modified: | 2016-01-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Latency and Function of Immune Inhibitor A Metallopeptidase, a Modulator of the Bacillus anthracis Secretome.
Structure, 24, 2016
|
|
6FK0
 
 | |
1PSM
 
 | |
1KZW
 
 | | Solution structure of Human Intestinal Fatty acid binding protein | | Descriptor: | INTESTINAL FATTY ACID-BINDING PROTEIN (A54) | | Authors: | Zhang, F, Luecke, C, Baier, L.J, Sacchettini, J.C, Hamilton, J.A. | | Deposit date: | 2002-02-08 | | Release date: | 2003-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human intestinal fatty acid binding protein with a naturally-occurring single amino acid substitution (A54T) that is associated with altered lipid metabolism
Biochemistry, 42, 2003
|
|
6FSN
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with UDP-glucose (conformation 1) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Roversi, P, Le Cornu, J.D, Hill, J, Alonzi, D.S, Zitzmann, N. | | Deposit date: | 2018-02-19 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Crystal polymorphism in fragment-based lead discovery of ligands of the catalytic domain of UGGT, the glycoprotein folding quality control checkpoint.
Front Mol Biosci, 2022
|
|
6GQ5
 
 | |
4WVM
 
 | | Stonustoxin structure | | Descriptor: | Stonustoxin subunit alpha, Stonustoxin subunit beta | | Authors: | Ellisdon, A.M, Panjikar, S, Whisstock, J.C, McGowan, S. | | Deposit date: | 2014-11-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Stonefish toxin defines an ancient branch of the perforin-like superfamily.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4X55
 
 | | Structure of the class D Beta-Lactamase OXA-225 K82D in Acyl-Enzyme Complex with Ceftazidime | | Descriptor: | 1-({(2R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-2-oxoethyl]-4-carboxy-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, Beta-lactamase OXA-225 | | Authors: | Clasman, J.R, June, C.M, Powers, R.A, Leonard, D.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.941 Å) | | Cite: | Structural Basis of Activity against Aztreonam and Extended Spectrum Cephalosporins for Two Carbapenem-Hydrolyzing Class D beta-Lactamases from Acinetobacter baumannii.
Biochemistry, 54, 2015
|
|
4YJY
 
 | |
6GQ2
 
 | |
6GQC
 
 | |
4YU6
 
 | | Crystal structure of Bacillus anthracis immune inhibitor A2 peptidase zymogen | | Descriptor: | ACETONITRILE, CALCIUM ION, Immune inhibitor A, ... | | Authors: | Arolas, J.L, Goulas, T, Gomis-Ruth, F.X. | | Deposit date: | 2015-03-18 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Latency and Function of Immune Inhibitor A Metallopeptidase, a Modulator of the Bacillus anthracis Secretome.
Structure, 24, 2016
|
|
1QDM
 
 | | CRYSTAL STRUCTURE OF PROPHYTEPSIN, A ZYMOGEN OF A BARLEY VACUOLAR ASPARTIC PROTEINASE. | | Descriptor: | PROPHYTEPSIN | | Authors: | Kervinen, J, Tobin, G.J, Costa, J, Waugh, D.S, Wlodawer, A, Zdanov, A. | | Deposit date: | 1999-05-19 | | Release date: | 1999-07-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of plant aspartic proteinase prophytepsin: inactivation and vacuolar targeting.
EMBO J., 18, 1999
|
|
7Z1H
 
 | | VAR2CSA APO | | Descriptor: | VAR2CSA APO | | Authors: | Raghavan, S.S.R, Wang, K.T. | | Deposit date: | 2022-02-24 | | Release date: | 2022-11-02 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM reveals the conformational epitope of human monoclonal antibody PAM1.4 broadly reacting with polymorphic malarial protein VAR2CSA.
Plos Pathog., 18, 2022
|
|
7Z12
 
 | | VAR2 complex with PAM1.4 | | Descriptor: | PAM1.4, Heavy Chain, light Chain, ... | | Authors: | Raghavan, S.S.R, Wang, K.T. | | Deposit date: | 2022-02-24 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM reveals the conformational epitope of human monoclonal antibody PAM1.4 broadly reacting with polymorphic malarial protein VAR2CSA.
Plos Pathog., 18, 2022
|
|
7K4A
 
 | | Cryo-EM structure of human TRPV6 in the open state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
6O5S
 
 | |
6URV
 
 | | Crystal structure of Yellow Fever Virus NS2B-NS3 protease domain | | Descriptor: | NS2B, NS3 protease | | Authors: | Noske, G.D, Gawriljuk, V.F.O, Fernandes, R.S, Oliva, G, Godoy, A.S. | | Deposit date: | 2019-10-24 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural characterization and polymorphism analysis of the NS2B-NS3 protease from the 2017 Brazilian circulating strain of Yellow Fever virus.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6MGL
 
 | | Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, D60A mutant in complex with XXLG and XGXXLG xyloglucan | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Arnal, G, Watanabe, N, Brumer, H, Savchenko, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural enzymology reveals the molecular basis of substrate regiospecificity and processivity of an exemplar bacterial glycoside hydrolase family 74endo-xyloglucanase.
Biochem. J., 475, 2018
|
|
6MGJ
 
 | | Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Nocek, B, Arnal, G, Brumer, H, Savchenko, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural enzymology reveals the molecular basis of substrate regiospecificity and processivity of an exemplar bacterial glycoside hydrolase family 74endo-xyloglucanase.
Biochem. J., 475, 2018
|
|
7KWE
 
 | | Crystal structure of the catalytic domain of human PDE3A bound to DNMDP | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Horner, S.W, Garvie, C. | | Deposit date: | 2020-11-30 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
6MGK
 
 | | Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, in complex with XLX xyloglucan | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Nocek, B, Arnal, G, Brumer, H, Savchenko, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural enzymology reveals the molecular basis of substrate regiospecificity and processivity of an exemplar bacterial glycoside hydrolase family 74endo-xyloglucanase.
Biochem. J., 475, 2018
|
|
