4LZ8
 
 | |
8CRI
 
 | | Crystal structure of LplA1 in complex with lipoic acid (Listeria monocytogenes) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, LIPOIC ACID, ... | | Authors: | Dienemann, J.-N, Chen, S.-Y, Hitzenberger, M, Sievert, M.L, Hacker, S.M, Prigge, S.T, Zacharias, M, Groll, M, Sieber, S.A. | | Deposit date: | 2023-03-08 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Chemical Proteomic Strategy Reveals Inhibitors of Lipoate Salvage in Bacteria and Parasites.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8D2T
 
 | | Zebrafish MFSD2A isoform B in inward open ligand-free conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
4UU5
 
 | | CRYSTAL STRUCTURE OF THE PDZ DOMAIN OF PALS1 IN COMPLEX WITH THE CRB PEPTIDE | | Descriptor: | 2,2,4,4,6,6,8-heptamethylnonane, CHLORIDE ION, MAGUK P55 SUBFAMILY MEMBER 5, ... | | Authors: | Ivanova, M.E, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2014-07-24 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structures of the human Pals1 PDZ domain with and without ligand suggest gated access of Crb to the PDZ peptide-binding groove.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
5WS5
 
 | | Native XFEL structure of photosystem II (preflash dark dataset) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Light-induced structural changes and the site of O=O bond formation in PSII caught by XFEL.
Nature, 543, 2017
|
|
5EIJ
 
 | | Carbonic Anhydrase II in complex with Sulfonamide Inhibitor | | Descriptor: | 1-(3-iodanylphenyl)-3-(4-sulfamoylphenyl)thiourea, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Lomelino, C.L, Mahon, B.P, McKenna, R. | | Deposit date: | 2015-10-29 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Kinetic and X-ray crystallographic investigations on carbonic anhydrase isoforms I, II, IX and XII of a thioureido analog of SLC-0111.
Bioorg. Med. Chem., 24, 2016
|
|
1B58
 
 | |
5ES3
 
 | | Co-crystal structure of LDH liganded with oxamate | | Descriptor: | L-lactate dehydrogenase A chain, OXAMIC ACID | | Authors: | Nowicki, M.W, Wear, M.A, McNae, I.W, Blackburn, E.A. | | Deposit date: | 2015-11-16 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Streamlined, Automated Protocol for the Production of Milligram Quantities of Untagged Recombinant Rat Lactate Dehydrogenase A Using AKTAxpressTM.
Plos One, 10, 2015
|
|
1B3G
 
 | |
1B4Z
 
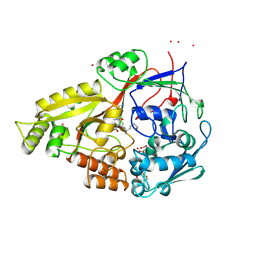 | | OLIGO-PEPTIDE BINDING PROTEIN (OPPA) COMPLEXED WITH KDK | | Descriptor: | ACETATE ION, PROTEIN (OLIGO-PEPTIDE BINDING PROTEIN), PROTEIN (PEPTIDE LYS-ASP-LYS), ... | | Authors: | Tame, J.R.H, Sleigh, S.H, Wilkinson, A.J. | | Deposit date: | 1999-01-04 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic and calorimetric analysis of peptide binding to OppA protein.
J.Mol.Biol., 291, 1999
|
|
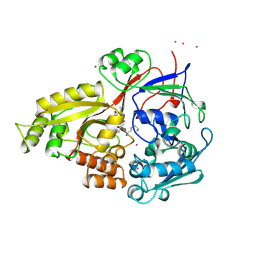
1B5J
 
 | |
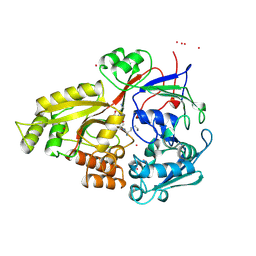
1B51
 
 | |
2VSR
 
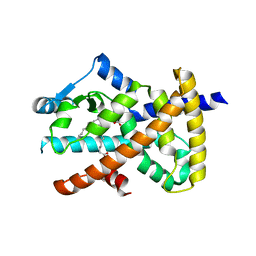 | | hPPARgamma Ligand binding domain in complex with 9-(S)-HODE | | Descriptor: | (9S,10E,12Z)-9-hydroxyoctadeca-10,12-dienoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-04-29 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
6WCQ
 
 | | Structure of a substrate-bound DQC ubiquitin ligase | | Descriptor: | Cullin-1, F-box/LRR-repeat protein 17, Kelch-like ECH-associated protein 1, ... | | Authors: | Mena, E.L, Jevtic, P, Greber, B.J, Gee, C.L, Lew, B.G, Akopian, D, Nogales, E, Kuriyan, J, Rape, M. | | Deposit date: | 2020-03-31 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Structural basis for dimerization quality control.
Nature, 586, 2020
|
|
2VST
 
 | | hPPARgamma Ligand binding domain in complex with 13-(S)-HODE | | Descriptor: | (9Z,11E,13S)-13-hydroxyoctadeca-9,11-dienoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-04-29 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV1
 
 | | hPPARgamma Ligand binding domain in complex with 4-HDHA | | Descriptor: | (4S,5E,7Z,10Z,13Z,16Z,19Z)-4-hydroxydocosa-5,7,10,13,16,19-hexaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
7BIT
 
 | | Inhibitor of MDM2-p53 Interaction | | Descriptor: | (3~{R})-2-[(5-chloranylpyridin-2-yl)methyl]-3-(4-chlorophenyl)-4-fluoranyl-3-[(1-oxidanylcyclopropyl)methoxy]-6-(2-oxidanylpropan-2-yl)isoindol-1-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Williams, P.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure-Based Design of Potent and Orally Active Isoindolinone Inhibitors of MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
2VV0
 
 | | hPPARgamma Ligand binding domain in complex with DHA | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV3
 
 | | hPPARgamma Ligand binding domain in complex with 4-oxoDHA | | Descriptor: | (6E,10Z,13Z,16Z,19Z)-4-oxodocosa-6,10,13,16,19-pentaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Activation of Ppargamma by Oxidized Fatty Acids.
Nat.Struct.Mol.Biol., 15, 2008
|
|
1B52
 
 | |
1B5I
 
 | |
2VV2
 
 | | hPPARgamma Ligand binding domain in complex with 5-HEPA | | Descriptor: | (5R,6E,8Z,11Z,14Z,17Z)-5-hydroxyicosa-6,8,11,14,17-pentaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV4
 
 | | hPPARgamma Ligand binding domain in complex with 6-oxoOTE | | Descriptor: | (8E,10S,12Z)-10-hydroxy-6-oxooctadeca-8,12-dienoic acid, (8R,9Z,12Z)-8-hydroxy-6-oxooctadeca-9,12-dienoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
6I94
 
 | |
4WRY
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-fluorouracil(B), Form I | | Descriptor: | 5-FLUOROURACIL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
