8REJ
 
 | | Crystal structure of PPAR gamma Ligand Binding Domain in complex with the ligand LBB78 | | Descriptor: | (2~{S})-2-(4-naphthalen-1-ylphenoxy)-3-phenyl-propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Capelli, D, Montanari, R. | | Deposit date: | 2023-12-11 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | A chemical modification of a peroxisome proliferator-activated receptor pan agonist produced a shift to a new dual alpha/gamma partial agonist endowed with mitochondrial pyruvate carrier inhibition and antidiabetic properties.
Eur.J.Med.Chem., 275, 2024
|
|
8RCE
 
 | | Crystal structure of PPAR alfa Ligand Binding Domain in complex with the ligand LBB78 | | Descriptor: | (2~{S})-2-(4-naphthalen-1-ylphenoxy)-3-phenyl-propanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Capelli, D, Montanari, R. | | Deposit date: | 2023-12-06 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A chemical modification of a peroxisome proliferator-activated receptor pan agonist produced a shift to a new dual alpha/gamma partial agonist endowed with mitochondrial pyruvate carrier inhibition and antidiabetic properties.
Eur.J.Med.Chem., 275, 2024
|
|
6R85
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-glutamate | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, Glutamate receptor 3.3,Glutamate receptor 3.3, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-03-31 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BJ6
 
 | | Inhibitor of MDM2-p53 Interaction | | Descriptor: | (3~{R})-2-[(5-chloranylpyridin-2-yl)methyl]-3-(4-chlorophenyl)-4-fluoranyl-3-[[1-(hydroxymethyl)cyclopropyl]methoxy]-6-(2-oxidanylpropan-2-yl)isoindol-1-one, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Williams, P.A. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-Based Design of Potent and Orally Active Isoindolinone Inhibitors of MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7BJ0
 
 | | Inhibitor of MDM2-p53 Interaction | | Descriptor: | (3~{R})-4-chloranyl-3-(4-chlorophenyl)-3-[[1-(hydroxymethyl)cyclopropyl]methoxy]-2-[(4-nitrophenyl)methyl]isoindol-1-one, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Williams, P.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of Potent and Orally Active Isoindolinone Inhibitors of MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7BMG
 
 | | Inhibitor of MDM2-p53 Interaction | | Descriptor: | (3~{R})-3-(4-chlorophenyl)-2-[(4-ethynylphenyl)methyl]-3-[[1-(hydroxymethyl)cyclopropyl]methoxy]-6-(2-oxidanylpropan-2-yl)isoindol-1-one, 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Williams, P.A. | | Deposit date: | 2021-01-20 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure-Based Design of Potent and Orally Active Isoindolinone Inhibitors of MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
6R8A
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-methionine | | Descriptor: | Glutamate receptor 3.3,Glutamate receptor 3.3, METHIONINE, SODIUM ION, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BIV
 
 | | Inhibitor of MDM2-p53 Interaction | | Descriptor: | 1,2-ETHANEDIOL, 6-[[(1~{R})-1-(4-chlorophenyl)-7-fluoranyl-1-[[1-(hydroxymethyl)cyclopropyl]methoxy]-3-oxidanylidene-5-(2-oxidanylpropan-2-yl)isoindol-2-yl]methyl]pyridine-3-carbonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Williams, P.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure-Based Design of Potent and Orally Active Isoindolinone Inhibitors of MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7BIR
 
 | | Inhibitor of MDM2-p53 Interaction | | Descriptor: | 1-[[(1~{R})-2-[(5-chloranylpyridin-2-yl)methyl]-1-(4-chlorophenyl)-7-fluoranyl-3-oxidanylidene-5-(2-oxidanylpropan-2-yl)isoindol-1-yl]oxymethyl]cyclopropane-1-carboxamide, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Williams, P.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-Based Design of Potent and Orally Active Isoindolinone Inhibitors of MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
4UU5
 
 | | CRYSTAL STRUCTURE OF THE PDZ DOMAIN OF PALS1 IN COMPLEX WITH THE CRB PEPTIDE | | Descriptor: | 2,2,4,4,6,6,8-heptamethylnonane, CHLORIDE ION, MAGUK P55 SUBFAMILY MEMBER 5, ... | | Authors: | Ivanova, M.E, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2014-07-24 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structures of the human Pals1 PDZ domain with and without ligand suggest gated access of Crb to the PDZ peptide-binding groove.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
6R88
 
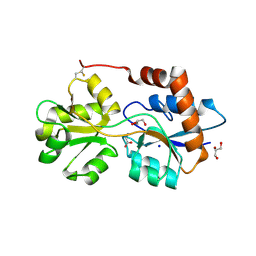 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with glycine | | Descriptor: | CHLORIDE ION, GLYCEROL, GLYCINE, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2ZNO
 
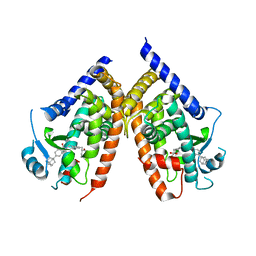 | | Human PPAR gamma ligand binding domain in complex with a synthetic agonist TIPP703 | | Descriptor: | (2S)-2-(4-propoxy-3-{[({4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]phenyl}carbonyl)amino]methyl}benzyl)butanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Oyama, T, Waku, T, Kasuga, J, Miyachi, H, Morikawa, K. | | Deposit date: | 2008-04-30 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Adaptability and selectivity of human peroxisome proliferator-activated receptor (PPAR) pan agonists revealed from crystal structures
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6R89
 
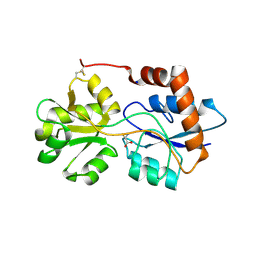 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-cysteine | | Descriptor: | CHLORIDE ION, CYSTEINE, GLYCEROL, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2VV0
 
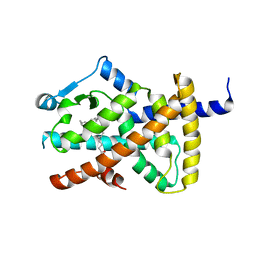 | | hPPARgamma Ligand binding domain in complex with DHA | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2ZNN
 
 | | Human PPAR alpha ligand binding domain in complex with a synthetic agonist TIPP703 | | Descriptor: | (2S)-2-(4-propoxy-3-{[({4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]phenyl}carbonyl)amino]methyl}benzyl)butanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Oyama, T, Toyota, K, Kasuga, J, Miyachi, H, Morikawa, K. | | Deposit date: | 2008-04-30 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Adaptability and selectivity of human peroxisome proliferator-activated receptor (PPAR) pan agonists revealed from crystal structures
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2VV2
 
 | | hPPARgamma Ligand binding domain in complex with 5-HEPA | | Descriptor: | (5R,6E,8Z,11Z,14Z,17Z)-5-hydroxyicosa-6,8,11,14,17-pentaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VSR
 
 | | hPPARgamma Ligand binding domain in complex with 9-(S)-HODE | | Descriptor: | (9S,10E,12Z)-9-hydroxyoctadeca-10,12-dienoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-04-29 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VST
 
 | | hPPARgamma Ligand binding domain in complex with 13-(S)-HODE | | Descriptor: | (9Z,11E,13S)-13-hydroxyoctadeca-9,11-dienoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-04-29 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV3
 
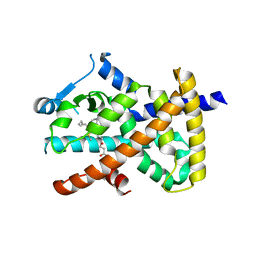 | | hPPARgamma Ligand binding domain in complex with 4-oxoDHA | | Descriptor: | (6E,10Z,13Z,16Z,19Z)-4-oxodocosa-6,10,13,16,19-pentaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Activation of Ppargamma by Oxidized Fatty Acids.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VV1
 
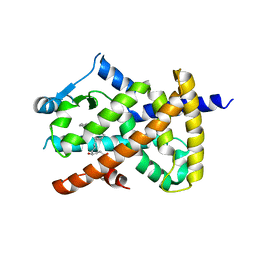 | | hPPARgamma Ligand binding domain in complex with 4-HDHA | | Descriptor: | (4S,5E,7Z,10Z,13Z,16Z,19Z)-4-hydroxydocosa-5,7,10,13,16,19-hexaenoic acid, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for the Activation of Pparg by Oxidised Fatty Acids
Nat.Struct.Mol.Biol., 15, 2008
|
|
2ZNQ
 
 | | Human PPAR delta ligand binding domain in complex with a synthetic agonist TIPP401 | | Descriptor: | (2S)-2-{3-[({[2-fluoro-4-(trifluoromethyl)phenyl]carbonyl}amino)methyl]-4-methoxybenzyl}butanoic acid, Peroxisome proliferator-activated receptor delta, heptyl beta-D-glucopyranoside | | Authors: | Oyama, T, Hirakawa, Y, Nagasawa, N, Miyachi, H, Morikawa, K. | | Deposit date: | 2008-04-30 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Adaptability and selectivity of human peroxisome proliferator-activated receptor (PPAR) pan agonists revealed from crystal structures
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2ZNP
 
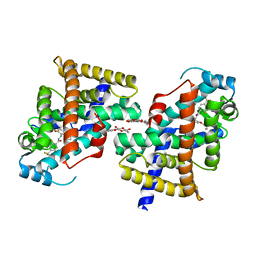 | | Human PPAR delta ligand binding domain in complex with a synthetic agonist TIPP204 | | Descriptor: | (2S)-2-{4-butoxy-3-[({[2-fluoro-4-(trifluoromethyl)phenyl]carbonyl}amino)methyl]benzyl}butanoic acid, Peroxisome proliferator-activated receptor delta, heptyl beta-D-glucopyranoside | | Authors: | Oyama, T, Hirakawa, Y, Nagasawa, N, Miyachi, H, Morikawa, K. | | Deposit date: | 2008-04-30 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Adaptability and selectivity of human peroxisome proliferator-activated receptor (PPAR) pan agonists revealed from crystal structures
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6VR7
 
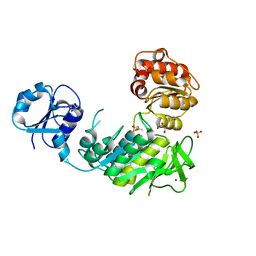 | | Structure of a pseudomurein peptide ligase type C from Methanothermus fervidus | | Descriptor: | ACETOACETIC ACID, GLYCEROL, Mur ligase middle domain protein, ... | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Archaeal pseudomurein and bacterial murein cell wall biosynthesis share a common evolutionary ancestry
FEMS Microbes, 2, 2021
|
|
4OER
 
 | | Crystal structure of NikA from Brucella suis, unliganded form | | Descriptor: | GLYCEROL, NikA protein, SULFATE ION | | Authors: | Lebrette, H, Cavazza, C. | | Deposit date: | 2014-01-13 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Promiscuous nickel import in human pathogens: structure, thermodynamics, and evolution of extracytoplasmic nickel-binding proteins.
Structure, 22, 2014
|
|
4OFO
 
 | | Crystal structure of YntA from Yersinia pestis, unliganded form | | Descriptor: | Extracytoplasmic Nickel-Binding Protein YpYntA, NICKEL (II) ION | | Authors: | Lebrette, H, Cavazza, C. | | Deposit date: | 2014-01-15 | | Release date: | 2014-10-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Promiscuous nickel import in human pathogens: structure, thermodynamics, and evolution of extracytoplasmic nickel-binding proteins.
Structure, 22, 2014
|
|
