6CP9
 
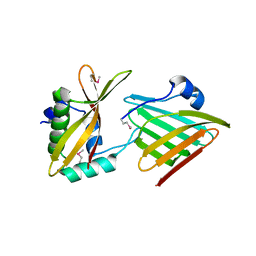 | | Contact-dependent growth inhibition toxin - immunity protein complex from Klebsiella pneumoniae 342 | | Descriptor: | CdiA, CdiI | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Convergent Evolution of the Barnase/EndoU/Colicin/RelE (BECR) Fold in Antibacterial tRNase Toxins.
Structure, 27, 2019
|
|
5C73
 
 | |
5C78
 
 | |
1KZX
 
 | | Solution structure of human intestinal fatty acid binding protein with a naturally-occurring single amino acid substitution (A54T) | | Descriptor: | INTESTINAL FATTY ACID-BINDING PROTEIN (T54) | | Authors: | Zhang, F, Luecke, C, Baier, L.J, Sacchettini, J.C, Hamilton, J.A. | | Deposit date: | 2002-02-08 | | Release date: | 2003-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human intestinal fatty acid binding protein with a naturally-occurring single amino acid substitution (A54T) that is associated with altered lipid metabolism
Biochemistry, 42, 2003
|
|
6CP8
 
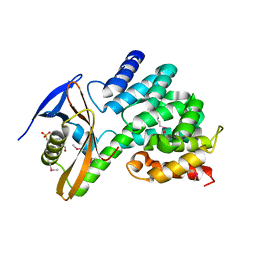 | | Contact-dependent growth inhibition toxin-immunity protein complex from from E. coli 3006 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CdiA, CdiI, ... | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Convergent Evolution of the Barnase/EndoU/Colicin/RelE (BECR) Fold in Antibacterial tRNase Toxins.
Structure, 27, 2019
|
|
7ZVA
 
 | | Crystal Structure of the native zymogen form of the glutamic-class prolyl-endopeptidase neprosin at 1.80 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, C-terminal peptidase, ... | | Authors: | Del Amo-Maestro, L, Eckhard, U, Rodriguez-Banqueri, A, Mendes, S.R, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular and in vivo studies of a glutamate-class prolyl-endopeptidase for coeliac disease therapy.
Nat Commun, 13, 2022
|
|
7ZU8
 
 | | Crystal Structure of the zymogen form of the glutamic-class prolyl-endopeptidase neprosin at 2.05 A resolution in presence of the crystallophore Lu-Xo4. | | Descriptor: | 12-oxidanyl-9,11$l^{3}-dioxa-1$l^{4},19$l^{4},22,27$l^{4},28$l^{4}-pentaza-10$l^{6}-lutetaoctacyclo[17.5.2.1^{3,7}.1^{10,13}.0^{1,10}.0^{10,19}.0^{10,28}.0^{17,27}]octacosa-3,5,7(28),11,13,15,17(27)-heptaen-8-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Del Amo-Maestro, L, Eckhard, U, Rodriguez-Banqueri, A, Mendes, S.R, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2022-05-11 | | Release date: | 2022-08-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular and in vivo studies of a glutamate-class prolyl-endopeptidase for coeliac disease therapy.
Nat Commun, 13, 2022
|
|
5DUO
 
 | | Crystal structure of native translocator protein 18kDa (TSPO) from Rhodobacter sphaeroides (A139T Mutant) in C2 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, FORMIC ACID, PROTOPORPHYRIN IX, ... | | Authors: | Li, F, Liu, J, Zheng, Y, Garavito, R.M, Ferguson-Miller, S. | | Deposit date: | 2015-09-20 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Response to Comment on "Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism".
Science, 350, 2015
|
|
5BTQ
 
 | | Crystal structure of human heme oxygenase 1 H25R with biliverdin bound | | Descriptor: | BILIVERDINE IX ALPHA, Heme oxygenase 1, SULFATE ION | | Authors: | Caaveiro, J.M.M, Morante, K, Sigala, P, Tsumoto, K. | | Deposit date: | 2015-06-03 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | In-Cell Enzymology To Probe His-Heme Ligation in Heme Oxygenase Catalysis
Biochemistry, 55, 2016
|
|
5E5J
 
 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 6.0 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Kovalevsky, A.Y, Gerlits, O.O. | | Deposit date: | 2015-10-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
1LW6
 
 | |
3IKY
 
 | | Structural model of ParM filament in the open state by cryo-EM | | Descriptor: | Plasmid segregation protein parM | | Authors: | Galkin, V.E, Orlova, A, Rivera, C, Mullins, R.D, Egelman, E.H. | | Deposit date: | 2009-08-06 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structural polymorphism of the ParM filament and dynamic instability
Structure, 17, 2009
|
|
3IKU
 
 | | Structural model of ParM filament in closed state from cryo-EM | | Descriptor: | Plasmid segregation protein parM | | Authors: | Galkin, V.E, Orlova, A, Rivera, C, Mullins, R.D, Egelman, E.H. | | Deposit date: | 2009-08-06 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structural polymorphism of the ParM filament and dynamic instability
Structure, 17, 2009
|
|
4IIS
 
 | | Crystal structure of a glycosylated beta-1,3-glucanase (HEV B 2), An allergen from Hevea Brasiliensis (Space group P41) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-1,3-glucanase form 'RRII Gln 2', CACODYLATE ION, ... | | Authors: | Rodriguez-Romero, A, Hernandez-Santoyo, A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6676 Å) | | Cite: | Structural analysis of the endogenous glycoallergen Hev b 2 (endo-beta-1,3-glucanase) from Hevea brasiliensis and its recognition by human basophils.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2AIH
 
 | | 1H-NMR solution structure of a trypsin/chymotrypsin Bowman-Birk inhibitor from Lens culinaris. | | Descriptor: | Bowman-Birk type protease inhibitor, LCTI, CHLORIDE ION | | Authors: | Ragg, E.M, Galbusera, V, Scarafoni, A, Negri, A, Tedeschi, G, Consonni, A, Sessa, F, Duranti, M. | | Deposit date: | 2005-07-29 | | Release date: | 2006-08-01 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Inhibitory properties and solution structure of a potent Bowman-Birk protease inhibitor from lentil (Lens culinaris, L) seeds.
Febs J., 273, 2006
|
|
4I04
 
 | | Structure of zymogen of cathepsin B1 from Schistosoma mansoni | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin B-like peptidase (C01 family) | | Authors: | Rezacova, P, Jilkova, A, Brynda, J, Horn, M, Mares, M. | | Deposit date: | 2012-11-16 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Activation route of the Schistosoma mansoni cathepsin B1 drug target: structural map with a glycosaminoglycan switch
Structure, 22, 2014
|
|
3VY6
 
 | |
3QVC
 
 | |
3VZF
 
 | |
3VZG
 
 | |
3VY7
 
 | |
6TL2
 
 | |
6SV2
 
 | |
6SUZ
 
 | |
5OI8
 
 | |
