7W9M
 
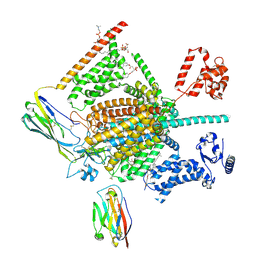 | | Cryo-EM structure of human Nav1.7(E406K) in complex with auxiliary beta subunits, ProTx-II and tetrodotoxin (S6IV pi helix conformer) | | Descriptor: | (1R,5R,6R,7R,9S,11S,12S,13S,14S)-3-amino-14-(hydroxymethyl)-8,10-dioxa-2,4-diazatetracyclo[7.3.1.1~7,11~.0~1,6~]tetradec-3-ene-5,9,12,13,14-pentol (non-preferred name), 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7W9P
 
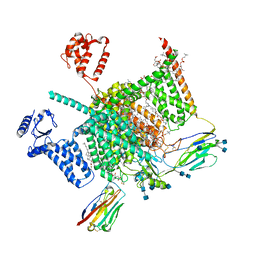 | | Cryo-EM structure of human Nav1.7(E406K) in complex with auxiliary beta subunits, huwentoxin-IV and saxitoxin (S6IV pi helix conformer) | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7W9T
 
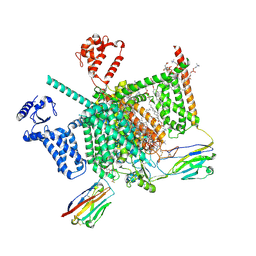 | | Cryo-EM structure of human Nav1.7(E406K) in complex with auxiliary beta subunits, huwentoxin-IV and saxitoxin (S6IV alpha helix conformer) | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7W9L
 
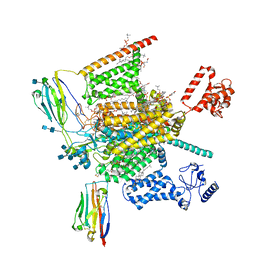 | | Cryo-EM structure of human Nav1.7(E406K)-beta1-beta2 complex | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | Deposit date: | 2021-12-10 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7WI0
 
 | | SARS-CoV-2 Omicron variant spike in complex with three human neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | Authors: | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
7WHZ
 
 | | SARS-CoV-2 spike protein in complex with three human neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | Authors: | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
7TJ8
 
 | | Cryo-EM structure of the human Nax channel in complex with beta3 solved in nanodiscs | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Noland, C.L, Kschonsak, M, Ciferri, C, Payandeh, J. | | Deposit date: | 2022-01-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-guided unlocking of Na X reveals a non-selective tetrodotoxin-sensitive cation channel.
Nat Commun, 13, 2022
|
|
7TJ9
 
 | | Cryo-EM structure of the human Nax channel in complex with beta3 solved in GDN | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Noland, C.L, Kschonsak, M, Ciferri, C, Payandeh, J. | | Deposit date: | 2022-01-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-guided unlocking of Na X reveals a non-selective tetrodotoxin-sensitive cation channel.
Nat Commun, 13, 2022
|
|
7QU6
 
 | | Crystal structure of the N-terminal domain of Siglec-8 | | Descriptor: | Sialic acid-binding Ig-like lectin 8 | | Authors: | Lenza, M.P, Atxabal, U, Nycholat, C.M, Oyenarte, I, Paulson, J.C, Franconetti, A, Quintana, J.I, Unione, L, Delgado, S, Jimenez-Barbero, J, Ereno-Orbea, J. | | Deposit date: | 2022-01-17 | | Release date: | 2023-01-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structures of the Inhibitory Receptor Siglec-8 in Complex with a High-Affinity Sialoside Analogue and a Therapeutic Antibody.
Jacs Au, 3, 2023
|
|
7QUI
 
 | | Crystal structure of the N-terminal domain of Siglec-8 in complex with sulfonamide sialoside analogue | | Descriptor: | (2~{S},4~{S},5~{R},6~{R})-5-acetamido-2-[(2~{S},3~{R},4~{S},5~{S},6~{R})-2-[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-acetamido-2-(hydroxymethyl)-4,6-bis(oxidanyl)oxan-3-yl]oxy-3,5-bis(oxidanyl)-6-(sulfooxymethyl)oxan-4-yl]oxy-6-[(1~{R},2~{R})-3-(naphthalen-2-ylsulfonylamino)-1,2-bis(oxidanyl)propyl]-4-oxidanyl-oxane-2-carboxylic acid, Sialic acid-binding Ig-like lectin 8 | | Authors: | Lenza, M.P, Oyenarte, I, Atxabal, U, Nycholat, C, Franconetti, A, Quintana, J.I, Delgado, S, Unione, L, Paulson, J, Jimenez-Barbero, J, Ereno-Orbea, J. | | Deposit date: | 2022-01-18 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.352 Å) | | Cite: | Structures of the Inhibitory Receptor Siglec-8 in Complex with a High-Affinity Sialoside Analogue and a Therapeutic Antibody.
Jacs Au, 3, 2023
|
|
7QUH
 
 | | Siglec-8 in complex with therapeutic Fab AK002. | | Descriptor: | Sialic acid-binding Ig-like lectin 8, Sialic acid-binding immunoglobulin-type lectin | | Authors: | Lenza, M.P, Oyenarte, I, Jimenez Barbero, J, Ereno Orbea, J. | | Deposit date: | 2022-01-18 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.867 Å) | | Cite: | Structures of the Inhibitory Receptor Siglec-8 in Complex with a High-Affinity Sialoside Analogue and a Therapeutic Antibody.
Jacs Au, 3, 2023
|
|
7WP1
 
 | | Cryo-EM structure of SARS-CoV-2 Mu S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, VacW-209 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WP5
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spikeprotein S1, VacW-209 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WON
 
 | | Cryo-EM structure of SARS-CoV-2 S2P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, VacW-209 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WP2
 
 | | Cryo-EM structure of SARS-CoV-2 C.1.2 S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, VacW-209 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WP0
 
 | | Cryo-EM structure of SARS-CoV-2 Delta S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c335_light_IGLV1-40_IGLJ3, Spike protein S1, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7TPS
 
 | | Crystal structure of ALPN-202 (engineered CD80 vIgD) in complex with PD-L1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Demonte, D.W, Maurer, M.F, Akutsu, M, Kimbung, Y.R, Logan, D.T, Walse, B. | | Deposit date: | 2022-01-26 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The engineered CD80 variant fusion therapeutic davoceticept combines checkpoint antagonism with conditional CD28 costimulation for anti-tumor immunity.
Nat Commun, 13, 2022
|
|
7WSL
 
 | | PD-1 in complex with Dostarlimab | | Descriptor: | Programmed cell death protein 1, heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2022-01-30 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Molecular basis of PD-1 blockade by dostarlimab, the FDA-approved antibody for cancer immunotherapy.
Biochem.Biophys.Res.Commun., 599, 2022
|
|
7WSO
 
 | | Structure of a membrane protein G | | Descriptor: | B-cell antigen receptor complex-associated protein alpha chain, B-cell antigen receptor complex-associated protein beta chain, Immunoglobulin heavy constant gamma 1 | | Authors: | Ma, X, Zhu, Y, Chen, Y, Huang, Z. | | Deposit date: | 2022-01-30 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Cryo-EM structures of two human B cell receptor isotypes.
Science, 377, 2022
|
|
7WSP
 
 | | Structure of a membrane protein M | | Descriptor: | B-cell antigen receptor complex-associated protein alpha chain, B-cell antigen receptor complex-associated protein beta chain, Isoform 2 of Immunoglobulin heavy constant mu | | Authors: | Ma, X, Zhu, Y, Chen, Y, Huang, Z. | | Deposit date: | 2022-01-30 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | Structure of a membrane protein M
To Be Published
|
|
7WVM
 
 | | The complex structure of PD-1 and cemiplimab | | Descriptor: | Heavy Chain of Cemiplimab, Light Chain of Cemiplimab, Programmed cell death protein 1 | | Authors: | Lu, D, Xu, Z.P, Liu, K.F, Tan, S.G, Gao, G.F, Chai, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | PD-1 N58-Glycosylation-Dependent Binding of Monoclonal Antibody Cemiplimab for Immune Checkpoint Therapy.
Front Immunol, 13, 2022
|
|
7TY0
 
 | |
7XAD
 
 | | Crystal strucutre of PD-L1 and DBL2_02 designed protein binder | | Descriptor: | DBL2_02 binder, Programmed cell death 1 ligand 1 | | Authors: | Liu, K.F, Xu, Z.P, Han, P, Pacesa, M, Gao, G.F, Chai, Y, Tan, S.G. | | Deposit date: | 2022-03-17 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7XAE
 
 | | Crystal strucutre of PD-L1 and 3ONJA protein | | Descriptor: | 2IC6, Programmed cell death 1 ligand 1 | | Authors: | Liu, K.F, Xu, Z.P, Han, P, Gao, G.F, Chai, Y, Tan, S.G. | | Deposit date: | 2022-03-17 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | Crystal strucutre of PD-L1 and 2IC6 protein
To Be Published
|
|
7UMG
 
 | | Crystal structure of human CD8aa-MR1-Ac-6-FP complex | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, Major histocompatibility complex class I-related gene protein, ... | | Authors: | Awad, W, Rossjohn, J. | | Deposit date: | 2022-04-06 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CD8 coreceptor engagement of MR1 enhances antigen responsiveness by human MAIT and other MR1-reactive T cells.
J.Exp.Med., 219, 2022
|
|
