7ZA8
 
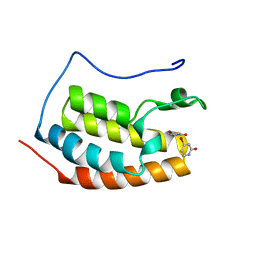 | | BRD4 in complex with FragLite6 | | Descriptor: | 4-bromanyl-1~{H}-pyridin-2-one, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-22 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7S9C
 
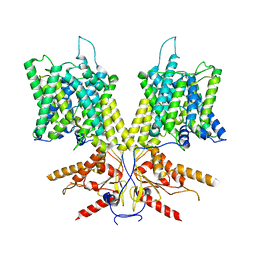 | | Cryo-EM Structure of dolphin Prestin: Sensor Down II (Expanded II) state | | Descriptor: | Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
7YPG
 
 | |
7ZCW
 
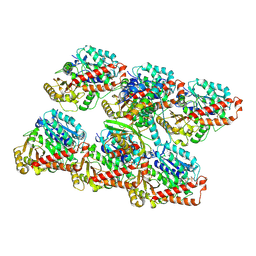 | | Cryo-EM structure of GMPCPP-microtubules in complex with VASH2-SVBP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Choi, S.R, Blum, T, Steinmetz, M.O. | | Deposit date: | 2022-03-29 | | Release date: | 2022-12-14 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | VASH1-SVBP and VASH2-SVBP generate different detyrosination profiles on microtubules.
J.Cell Biol., 222, 2023
|
|
7S9D
 
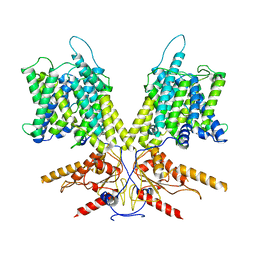 | | Cryo-EM Structure of dolphin Prestin: Intermediate state | | Descriptor: | Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
7Z07
 
 | |
7Z08
 
 | |
7Z4B
 
 | | Bacteriophage SU10 virion (C1) | | Descriptor: | Adaptor, Major head protein, Portal protein, ... | | Authors: | Siborova, M, Fuzik, T, Prochazkova, M, Novacek, J, Plevka, P. | | Deposit date: | 2022-03-03 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Tail proteins of phage SU10 reorganize into the nozzle for genome delivery.
Nat Commun, 13, 2022
|
|
7YV9
 
 | |
7Z0B
 
 | |
7S9A
 
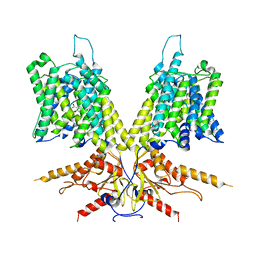 | | Cryo-EM Structure of dolphin Prestin: Inhibited I (Chloride + Salicylate) | | Descriptor: | 2-HYDROXYBENZOIC ACID, Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
7S9B
 
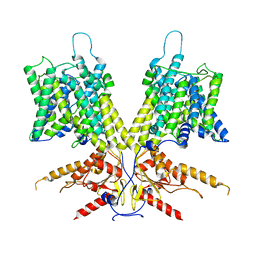 | | Cryo-EM Structure of dolphin Prestin: Sensor Down I (Expanded) state | | Descriptor: | Prestin | | Authors: | Bavi, N, Clark, M.D, Contreras, G.F, Shen, R, Reddy, B.G, Milewski, W, Perozo, E. | | Deposit date: | 2021-09-20 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The conformational cycle of prestin underlies outer-hair cell electromotility.
Nature, 600, 2021
|
|
7Z3S
 
 | |
7ZGB
 
 | | Structure of yeast Sec14p with NPPM112 | | Descriptor: | 4-fluoranyl-~{N}-[(4-pyrrolidin-1-ylphenyl)methyl]benzamide, SEC14 cytosolic factor | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2022-04-03 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanisms by which small molecules of diverse chemotypes arrest Sec14 lipid transfer activity.
J.Biol.Chem., 299, 2023
|
|
7Z62
 
 | | Structure of the LecA lectin from Pseudomonas aeruginosa in complex with a biaryl-thiogalactoside | | Descriptor: | (2R,3R,4S,5R,6S)-2-(hydroxymethyl)-6-[4-[1-(4-methoxyphenyl)ethenyl]phenyl]sulfanyl-oxane-3,4,5-triol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Varrot, A. | | Deposit date: | 2022-03-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovery of potent 1,1-diarylthiogalactoside glycomimetic inhibitors of Pseudomonas aeruginosa LecA with antibiofilm properties.
Eur.J.Med.Chem., 247, 2022
|
|
7YZT
 
 | | Crystal structure of a dye-decolorizing (Dyp) peroxidase from Acinetobacter radioresistens | | Descriptor: | Dyp-type peroxidase family protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhang, C, Catucci, G, Gilardi, G, Sadeghi, S.J, Di Nardo, G. | | Deposit date: | 2022-02-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a decolorizing peroxidase (Dyp) from Acinetobacter radioresistens
To Be Published
|
|
7YQ0
 
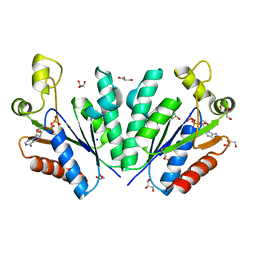 | | Crystal structure of adenosine 5'-phosphosulfate kinase from Archaeoglobus fulgidus in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenylyl-sulfate kinase, GLYCEROL, ... | | Authors: | Kawakami, T, Teramoto, T, Kakuta, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of adenosine 5'-phosphosulfate kinase isolated from Archaeoglobus fulgidus.
Biochem.Biophys.Res.Commun., 643, 2022
|
|
7YQ1
 
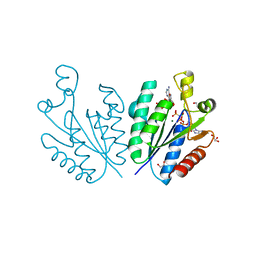 | | Crystal structure of adenosine 5'-phosphosulfate kinase from Archaeoglobus fulgidus in complex with AMP-PNP and APS | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, Adenylyl-sulfate kinase, BORIC ACID, ... | | Authors: | Kawakami, T, Teramoto, T, Kakuta, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of adenosine 5'-phosphosulfate kinase isolated from Archaeoglobus fulgidus.
Biochem.Biophys.Res.Commun., 643, 2022
|
|
7ZCH
 
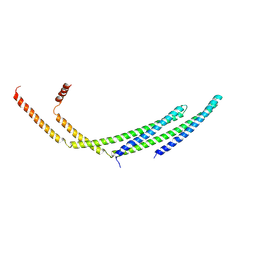 | | CHMP2A-CHMP3 heterodimer (410 Angstrom diameter) | | Descriptor: | Charged multivesicular body protein 2a, Charged multivesicular body protein 3 | | Authors: | Azad, K, Desfosses, A, Effantin, G, Schoehn, G, Weissenhorn, W. | | Deposit date: | 2022-03-28 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of CHMP2A-CHMP3 ESCRT-III polymer assembly and membrane cleavage.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YNY
 
 | |
7ZG9
 
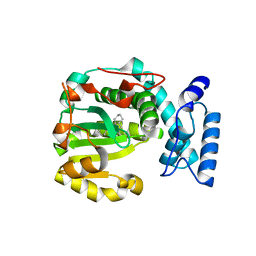 | | Structure of yeast Sec14p with himbacine | | Descriptor: | Himbacine, SEC14 cytosolic factor | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2022-04-03 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Mechanisms by which small molecules of diverse chemotypes arrest Sec14 lipid transfer activity.
J.Biol.Chem., 299, 2023
|
|
7YYO
 
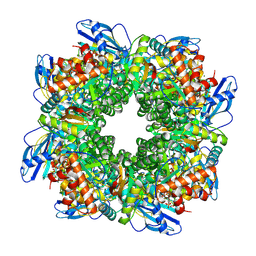 | | Cryo-EM structure of an a-carboxysome RuBisCO enzyme at 2.9 A resolution | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Mann, D, Evans, S.L, Bergeron, J.R.C. | | Deposit date: | 2022-02-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Single-particle cryo-EM analysis of the shell architecture and internal organization of an intact alpha-carboxysome.
Structure, 31, 2023
|
|
7ZGA
 
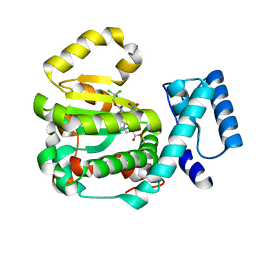 | | Structure of yeast Sec14p with ergoline | | Descriptor: | SEC14 cytosolic factor, ~{O}9-methyl ~{O}4-[2,2,2-tris(chloranyl)ethyl] (5~{a}~{S},6~{a}~{S},9~{R},10~{a}~{S})-7-methyl-3-nitro-5,5~{a},6,6~{a},8,9,10,10~{a}-octahydroindolo[4,3-fg]quinoline-4,9-dicarboxylate | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2022-04-03 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Mechanisms by which small molecules of diverse chemotypes arrest Sec14 lipid transfer activity.
J.Biol.Chem., 299, 2023
|
|
7ZGC
 
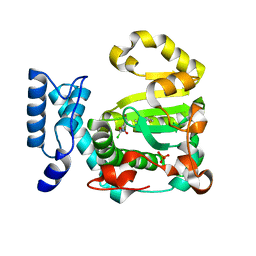 | | Structure of yeast Sec14p with NPPM481 | | Descriptor: | (4-chloranyl-3-nitro-phenyl)-[4-(2-fluorophenyl)piperazin-1-yl]methanone, PHOSPHATE ION, SEC14 cytosolic factor | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2022-04-03 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.236 Å) | | Cite: | Mechanisms by which small molecules of diverse chemotypes arrest Sec14 lipid transfer activity.
J.Biol.Chem., 299, 2023
|
|
7Z63
 
 | |
