1KXB
 
 | |
1KXE
 
 | |
4H6T
 
 | | Crystal structure of prethrombin-2 mutant E14eA/D14lA/E18A/S195A | | Descriptor: | PHOSPHATE ION, Prothrombin | | Authors: | Pozzi, N, Chen, Z, Zapata, F, Pelc, L.A, Di Cera, E. | | Deposit date: | 2012-09-19 | | Release date: | 2013-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Autoactivation of thrombin precursors.
J.Biol.Chem., 288, 2013
|
|
7X6R
 
 | | Crystal structure of actinomycin D-echinomycin-d(AGCACGT/ACGGGCT) complex | | Descriptor: | 2-CARBOXYQUINOXALINE, Actinomycin D, COBALT (II) ION, ... | | Authors: | Kao, S.H, Satange, R.B, Hou, M.H. | | Deposit date: | 2022-03-08 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Staggered intercalation of DNA duplexes with base-pair modulation by two distinct drug molecules induces asymmetric backbone twisting and structure polymorphism.
Nucleic Acids Res., 50, 2022
|
|
7X9F
 
 | | Crystal structure of actinomycin D-echinomycin-d(AGCGCGT/ACGCGCT) complex | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(P*AP*CP*GP*CP*GP*CP*T)-3'), DNA (5'-D(P*AP*GP*CP*GP*CP*GP*T)-3'), ... | | Authors: | Kao, S.H, Satange, R.B, Hou, M.H. | | Deposit date: | 2022-03-15 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Staggered intercalation of DNA duplexes with base-pair modulation by two distinct drug molecules induces asymmetric backbone twisting and structure polymorphism.
Nucleic Acids Res., 50, 2022
|
|
7X97
 
 | | Crystal structure of actinomycin D-echinomycin-d(AGCCCGT/ACGGGCT) complex | | Descriptor: | 2-CARBOXYQUINOXALINE, Actinomycin D, CHLORIDE ION, ... | | Authors: | Kao, S.H, Satange, R.B, Hou, M.H. | | Deposit date: | 2022-03-15 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Staggered intercalation of DNA duplexes with base-pair modulation by two distinct drug molecules induces asymmetric backbone twisting and structure polymorphism.
Nucleic Acids Res., 50, 2022
|
|
7XDJ
 
 | | Crystal structure of actinomycin D-echinomycin-d(AGCGCGT/ACGAGCT) complex | | Descriptor: | 2-CARBOXYQUINOXALINE, CHLORIDE ION, DNA (5'-D(P*AP*CP*GP*AP*GP*CP*(BRU))-3'), ... | | Authors: | Chien, C.M, Satange, R.B, Hou, M.H. | | Deposit date: | 2022-03-27 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.435 Å) | | Cite: | Staggered intercalation of DNA duplexes with base-pair modulation by two distinct drug molecules induces asymmetric backbone twisting and structure polymorphism.
Nucleic Acids Res., 50, 2022
|
|
5FFN
 
 | | Complex of subtilase SubTY from Bacillus sp. TY145 with chymotrypsin inhibitor CI2A | | Descriptor: | CALCIUM ION, Enzyme subtilase SubTY from Bacillus sp. TY145, SODIUM ION, ... | | Authors: | McAuley, K.E, Svendsen, A, Oestergaard, P.R, Dohnalek, J, Wilson, K.S. | | Deposit date: | 2015-12-18 | | Release date: | 2016-05-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stabilization of Enzymes by Metal Binding: Structures of Two Alkalophilic Bacillus Subtilases and Analysis of the Second Metal-Binding Site of the Subtilase Family
Book, 2016
|
|
1ESC
 
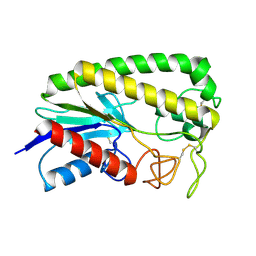 | | THE MOLECULAR MECHANISM OF ENANTIORECOGNITION BY ESTERASES | | Descriptor: | ESTERASE | | Authors: | Wei, Y, Schottel, J.L, Derewenda, U, Swenson, L, Patkar, S, Derewenda, Z.S. | | Deposit date: | 1994-10-07 | | Release date: | 1995-10-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel variant of the catalytic triad in the Streptomyces scabies esterase.
Nat.Struct.Biol., 2, 1995
|
|
9EWW
 
 | | Human p53 mRNA_CDS_120nt | | Descriptor: | p53 mRNA_CDS_120nt | | Authors: | Wang, L, Chen, S. | | Deposit date: | 2024-04-05 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (8.57 Å) | | Cite: | A cancer-derived synonymous mutation targets the DNA damage-activated p53 mRNA riboswitch.
To Be Published
|
|
9FKD
 
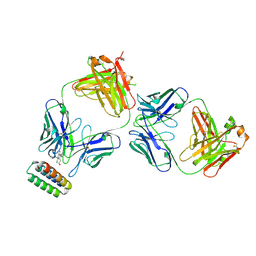 | | Progesterone-bound DB3 Fab in complex with computationally designed DBPro1156_2 protein binder | | Descriptor: | Anti-kappa Fab Heavy Chain, Anti-kappa Fab Light Chain, DB3 Fab Heavy chain, ... | | Authors: | Pacesa, M, Marchand, A, Correia, B.E. | | Deposit date: | 2024-06-03 | | Release date: | 2024-10-30 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Targeting protein-ligand neosurfaces with a generalizable deep learning tool.
Nature, 639, 2025
|
|
5SRR
 
 | |
5OPH
 
 | |
6ASC
 
 | | Mre11 dimer in complex with Endonuclease inhibitor PFM04 | | Descriptor: | (5E)-3-butyl-5-[(4-hydroxyphenyl)methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one, 1,2-ETHANEDIOL, MANGANESE (II) ION, ... | | Authors: | Moiani, D, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2017-08-24 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Targeting Allostery with Avatars to Design Inhibitors Assessed by Cell Activity: Dissecting MRE11 Endo- and Exonuclease Activities.
Meth. Enzymol., 601, 2018
|
|
5NKZ
 
 | | Crystal structure of H. polymorpha ubiquitin conjugating enzyme Pex4p in complex with soluble domain of Pex22p | | Descriptor: | Peroxin 22, Ubiquitin-conjugating enzyme E2-21 kDa | | Authors: | Danda, N, Lunev, S, Ali, A, Groves, M.R, Williams, C. | | Deposit date: | 2017-04-03 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into K48-linked ubiquitin chain formation by the Pex4p-Pex22p complex.
Biochem. Biophys. Res. Commun., 496, 2018
|
|
5O2D
 
 | | PARP14 Macrodomain 2 with inhibitor | | Descriptor: | Poly [ADP-ribose] polymerase 14, ~{N}-[2-(9~{H}-carbazol-1-yl)phenyl]methanesulfonamide | | Authors: | Uth, K, Schuller, M, Sieg, C, Wang, J, Krojer, T, Knapp, S, Riedels, K, Bracher, F, Edwards, A.M, Arrowsmith, C, Bountra, C, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-05-20 | | Release date: | 2017-11-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of a Selective Allosteric Inhibitor Targeting Macrodomain 2 of Polyadenosine-Diphosphate-Ribose Polymerase 14.
ACS Chem. Biol., 12, 2017
|
|
6FGE
 
 | | Crystal structure of human ZUFSP/ZUP1 in complex with ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, AMMONIUM ION, ... | | Authors: | Kwasna, D, Abdul Rehman, S.A, Kulathu, Y. | | Deposit date: | 2018-01-10 | | Release date: | 2018-04-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery and Characterization of ZUFSP/ZUP1, a Distinct Deubiquitinase Class Important for Genome Stability.
Mol. Cell, 70, 2018
|
|
1K9Y
 
 | | The PAPase Hal2p complexed with magnesium ions and reaction products: AMP and inorganic phosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-MERCAPTOETHANOL, Halotolerance protein HAL2, ... | | Authors: | Patel, S, Albert, A, Blundell, T.L. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural enzymology of Li(+)-sensitive/Mg(2+)-dependent phosphatases.
J.Mol.Biol., 320, 2002
|
|
1KA1
 
 | | The PAPase Hal2p complexed with calcium and magnesium ions and reaction substrate: PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Patel, S, Albert, A, Blundell, T.L. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural enzymology of Li(+)-sensitive/Mg(2+)-dependent phosphatases.
J.Mol.Biol., 320, 2002
|
|
4YPC
 
 | |
1VCP
 
 | |
1VCQ
 
 | |
5JOD
 
 | | Structure of proplasmepsin IV from Plasmodium falciparum | | Descriptor: | GLYCEROL, Proplasmepsin IV | | Authors: | Recacha, R, Akopjana, I, Tars, K, Jaudzems, K. | | Deposit date: | 2016-05-02 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | Crystal structure of Plasmodium falciparum proplasmepsin IV: the plasticity of proplasmepsins.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4YV3
 
 | |
1SSX
 
 | |
