4ZU9
 
 | | Crystal structure of bacterial selenocysteine-specific elongation factor EF-Sec | | Descriptor: | CYSTEINE, Elongation factor SelB, MAGNESIUM ION, ... | | Authors: | Itoh, Y, Sekine, S, Yokoyama, S. | | Deposit date: | 2015-05-15 | | Release date: | 2015-09-23 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (3.191 Å) | | Cite: | Crystal structure of the full-length bacterial selenocysteine-specific elongation factor SelB
Nucleic Acids Res., 43, 2015
|
|
1J53
 
 | | Structure of the N-terminal Exonuclease Domain of the Epsilon Subunit of E.coli DNA Polymerase III at pH 8.5 | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase III, epsilon chain, ... | | Authors: | Hamdan, S, Carr, P.D, Brown, S.E, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2002-01-22 | | Release date: | 2002-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Proofreading during Replication of the Escherichia coli Chromosome
Structure, 10, 2002
|
|
1VSE
 
 | | ASV INTEGRASE CORE DOMAIN WITH MG(II) COFACTOR AND HEPES LIGAND, LOW MG CONCENTRATION FORM | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, INTEGRASE | | Authors: | Bujacz, G, Jaskolski, M, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The catalytic domain of avian sarcoma virus integrase: conformation of the active-site residues in the presence of divalent cations.
Structure, 4, 1996
|
|
4V0L
 
 | |
1VUB
 
 | | CCDB, A TOPOISOMERASE POISON FROM E. COLI | | Descriptor: | CCDB, CHLORIDE ION | | Authors: | Loris, R, Dao-Thi, M.-H, Bahasi, E.M, Van Melderen, L, Poortmans, F, Liddington, R, Couturier, M, Wyns, L. | | Deposit date: | 1998-04-17 | | Release date: | 1998-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of CcdB, a topoisomerase poison from E. coli.
J.Mol.Biol., 285, 1999
|
|
1MS6
 
 | | Dipeptide Nitrile Inhibitor Bound to Cathepsin S. | | Descriptor: | Cathepsin S, MORPHOLINE-4-CARBOXYLIC ACID [1S-(2-BENZYLOXY-1R-CYANO-ETHYLCARBAMOYL)-3-METHYL-BUTYL]AMIDE | | Authors: | Ward, Y.D, Thomson, D.S, Frye, L.L, Cywin, C.L, Morwick, T, Emmanuel, M.J, Zindell, R, McNeil, D, Bekkali, Y, Giradot, M, Hrapchak, M, DeTuri, M, Crane, K, White, D, Pav, S, Wang, Y, Hao, M.H, Grygon, C.A, Labadia, M.E, Freeman, D.M, Davidson, W, Hopkins, J.L, Brown, M.L, Spero, D.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-04-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and synthesis of dipeptide nitriles as reversible and potent Cathepsin S inhibitors
J.Med.Chem., 45, 2002
|
|
6XNV
 
 | | CRYSTAL STRUCTURE OF LISTERIA MONOCYTOGENES CBPB PROTEIN (LMO1009) IN COMPLEX WITH C-DI-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CBS domain-containing protein | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2020-07-04 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | (p)ppGpp and c-di-AMP Homeostasis Is Controlled by CbpB in Listeria monocytogenes.
Mbio, 11, 2020
|
|
2DDS
 
 | | Crystal structure of sphingomyelinase from Bacillus cereus with cobalt ion | | Descriptor: | COBALT (II) ION, Sphingomyelin phosphodiesterase | | Authors: | Ago, H, Oda, M, Takahashi, M, Tsuge, H, Ochi, S, Katunuma, N, Miyano, M, Sakurai, J, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-02 | | Release date: | 2006-05-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of the Sphingomyelin Phosphodiesterase Activity in Neutral Sphingomyelinase from Bacillus cereus.
J.Biol.Chem., 281, 2006
|
|
5WC3
 
 | | SpoIIIAG | | Descriptor: | SpoIIIAG, Stage III sporulation engulfment assemblyprotein | | Authors: | Zeytuni, N, Hong, C, Worrall, L.J, Huang, R.K, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2017-06-29 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Near-atomic resolution cryoelectron microscopy structure of the 30-fold homooligomeric SpoIIIAG channel essential to spore formation in Bacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6XPB
 
 | |
4ZWT
 
 | |
5WEK
 
 | | GluA2 bound to antagonist ZK and GSG1L in digitonin, state 1 | | Descriptor: | Chimera of Glutamate receptor 2,Germ cell-specific gene 1-like protein, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2017-07-10 | | Release date: | 2017-08-02 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Channel opening and gating mechanism in AMPA-subtype glutamate receptors.
Nature, 549, 2017
|
|
1MTO
 
 | | Crystal structure of a Phosphofructokinase mutant from Bacillus stearothermophilus bound with fructose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructokinase | | Authors: | Riley-Lovingshimer, M.R, Ronning, D.R, Sacchettini, J.C, Reinhart, G.D. | | Deposit date: | 2002-09-21 | | Release date: | 2002-12-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Reversible Ligand-Induced Dissociation of a Tryptophan-Shift Mutant of Phosphofructokinase
from Bacillus stearothermophilus
Biochemistry, 41, 2002
|
|
7NE6
 
 | | Human TET2 in complex with unfavourable DNA substrate. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*AP*CP*AP*GP*GP*(5CM)P*GP*CP*CP*TP*G)-3'), ... | | Authors: | Rafalski, D, Bochtler, M. | | Deposit date: | 2021-02-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pronounced sequence specificity of the TET enzyme catalytic domain guides its cellular function.
Sci Adv, 8, 2022
|
|
3VBV
 
 | |
3VKZ
 
 | |
1VSF
 
 | | ASV INTEGRASE CORE DOMAIN WITH MN(II) COFACTOR AND HEPES LIGAND, HIGH MG CONCENTRATION FORM | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, INTEGRASE, MANGANESE (II) ION | | Authors: | Bujacz, G, Jaskolski, M, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The catalytic domain of avian sarcoma virus integrase: conformation of the active-site residues in the presence of divalent cations.
Structure, 4, 1996
|
|
6XU4
 
 | | Crystal structure of the genetically-encoded FGCaMP calcium indicator in its calcium-bound state | | Descriptor: | CALCIUM ION, FGCamp | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Barykina, N.V, Subach, O.M, Subach, F.V. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | FGCaMP7, an Improved Version of Fungi-Based Ratiometric Calcium Indicator for In Vivo Visualization of Neuronal Activity.
Int J Mol Sci, 21, 2020
|
|
5WKV
 
 | |
6RLV
 
 | | Trypanosoma brucei Seryl-tRNA Synthetase in Complex with 5'-O-(N-(L-seryl)-sulfamoyl)N3-methyluridine | | Descriptor: | 5'-O-(N-(L-seryl)-sulfamoyl)N3-methyluridine, GLYCEROL, MALONATE ION, ... | | Authors: | Pang, L, De Graef, S, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
6XXK
 
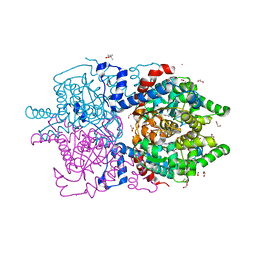 | | Crystal Structure of Human Deoxyhypusine Synthase in complex with spermidine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Wator, E, Wilk, P, Grudnik, P. | | Deposit date: | 2020-01-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Half Way to Hypusine-Structural Basis for Substrate Recognition by Human Deoxyhypusine Synthase.
Biomolecules, 10, 2020
|
|
5WPT
 
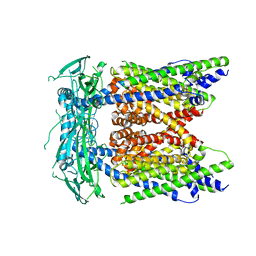 | | Cryo-EM structure of mammalian endolysosomal TRPML1 channel in nanodiscs in closed II conformation at 3.75 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-1, SODIUM ION | | Authors: | Chen, Q, She, J, Guo, J, Bai, X, Jiang, Y. | | Deposit date: | 2017-08-07 | | Release date: | 2017-10-18 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure of mammalian endolysosomal TRPML1 channel in nanodiscs.
Nature, 550, 2017
|
|
3VOM
 
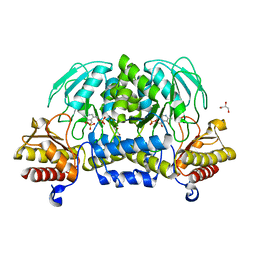 | | Structure of a putative phosphoserine aminotransferase from mycobacterium tuberculosis | | Descriptor: | GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, Putative phosphoserine aminotransferase, ... | | Authors: | Coulibaly, F, Lassalle, E, Baker, H.M, Baker, E.N. | | Deposit date: | 2012-01-30 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of phosphoserine aminotransferase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6RNN
 
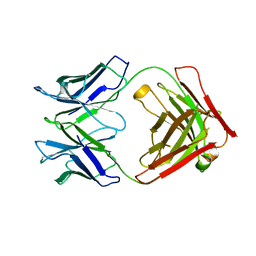 | | P46, an immunodominant surface protein from Mycoplasma hyopneumoniae | | Descriptor: | Immunoglobulin heavy chain, Immunoglobulin light chain | | Authors: | Guasch, A, Gonzalez-Gonzalez, L, Fita, I. | | Deposit date: | 2019-05-09 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of P46, an immunodominant surface protein from Mycoplasma hyopneumoniae: interaction with a monoclonal antibody.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6XJD
 
 | | Two mouse cGAS catalytic domain binding to human assembled nucleosome | | Descriptor: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1, ... | | Authors: | Xu, P, Li, P, Zhao, B. | | Deposit date: | 2020-06-23 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | The molecular basis of tight nuclear tethering and inactivation of cGAS.
Nature, 587, 2020
|
|
