7N6M
 
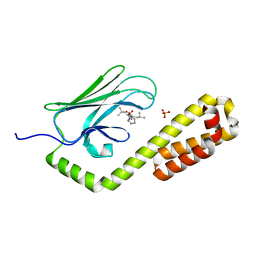 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide RQKPLLGLSR | | Descriptor: | Alkaline phosphatase peptide, Chaperone protein DnaK, SULFATE ION | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2021-06-08 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2NYB
 
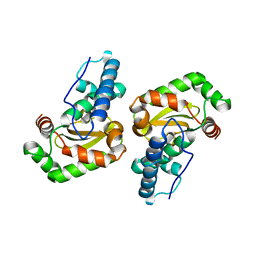 | |
7JMM
 
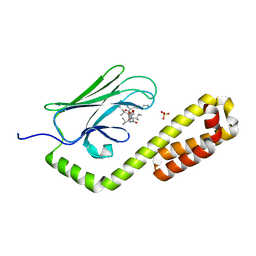 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide RAKNIILLSR | | Descriptor: | Alkaline phosphatase, Chaperone protein DnaK, SULFATE ION | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2020-08-02 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5CCC
 
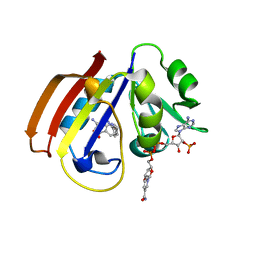 | |
7JNE
 
 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide RGSQLRIASR | | Descriptor: | Alkaline phosphatase peptide, Chaperone protein DnaK, SULFATE ION | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5CC9
 
 | |
5N2R
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with PSB36 at 2.8A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-butyl-3-[(~{E})-3-oxidanylprop-1-enyl]-8-[(1~{R},5~{S})-3-tricyclo[3.3.1.0^{3,7}]nonanyl]-7~{H}-purine-2,6-dione, Adenosine receptor A2a,Soluble cytochrome b562,ADENOSINE RECEPTOR A2A SOLUBLE CYTOCHROME B562 ADENOSINE RECEPTOR A2A,Adenosine receptor A2a, ... | | Authors: | Cheng, R.K.Y, Segala, E, Robertson, N, Deflorian, F, Dore, A.S, Errey, J.C, Fiez-Vandal, C, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2017-02-08 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity.
Structure, 25, 2017
|
|
1ZN3
 
 | | Crystal structure of Glu335Ala mutant of Clostridium botulinum neurotoxin type E | | Descriptor: | CHLORIDE ION, ZINC ION, botulinum neurotoxin type E | | Authors: | Agarwal, R, Binz, T, Swaminathan, S. | | Deposit date: | 2005-05-11 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Analysis of Active Site Residues of Botulinum Neurotoxin E by Mutational, Functional, and Structural Studies: Glu335Gln Is an Apoenzyme.
Biochemistry, 44, 2005
|
|
1D6K
 
 | | NMR SOLUTION STRUCTURE OF THE 5S RRNA E-LOOP/L25 COMPLEX | | Descriptor: | 5S RRNA E-LOOP (5SE), RIBOSOMAL PROTEIN L25 | | Authors: | Stoldt, M, Wohnert, J, Ohlenschlager, O, Gorlach, M, Brown, L.R. | | Deposit date: | 1999-10-14 | | Release date: | 1999-11-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the 5S rRNA E-domain-protein L25 complex shows preformed and induced recognition.
EMBO J., 18, 1999
|
|
2R6P
 
 | | Fit of E protein and Fab 1A1D-2 into 24 angstrom resolution cryoEM map of Fab complexed with dengue 2 virus. | | Descriptor: | Heavy chain of 1A1D-2, Light chain of 1A1D-2, Major envelope protein E | | Authors: | Lok, S.M, Kostyuchenko, V.K, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins.
Nat.Struct.Mol.Biol., 15, 2008
|
|
1GKS
 
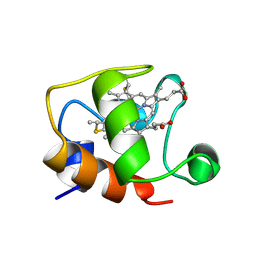 | | ECTOTHIORHODOSPIRA HALOPHILA CYTOCHROME C551 (REDUCED), NMR, 37 STRUCTURES | | Descriptor: | CYTOCHROME C551, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bersch, B, Blackledge, M.J, Meyer, T.E, Marion, D. | | Deposit date: | 1996-07-16 | | Release date: | 1997-01-27 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Ectothiorhodospira halophila ferrocytochrome c551: solution structure and comparison with bacterial cytochromes c.
J.Mol.Biol., 264, 1996
|
|
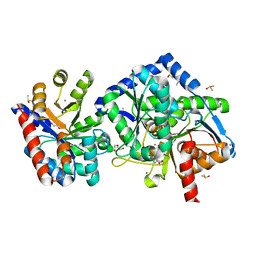
7KQ9
 
 | |
7BAR
 
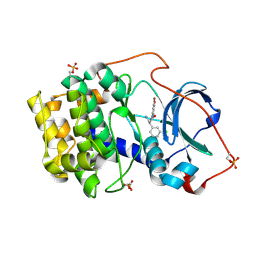 | |
3JZ1
 
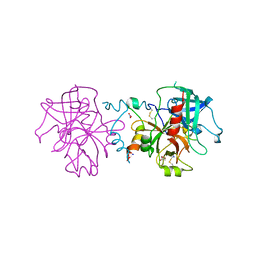 | | Crystal structure of human thrombin mutant N143P in E:Na+ form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, NITRATE ION, ... | | Authors: | Niu, W, Chen, Z, Bush-Pelc, L.A, Bah, A, Gandhi, P.S, Di Cera, E. | | Deposit date: | 2009-09-22 | | Release date: | 2009-10-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mutant N143P reveals how Na+ activates thrombin
J.Biol.Chem., 284, 2009
|
|
7LC6
 
 | | Cryo-EM Structure of KdpFABC in E2-P state with BeF3 | | Descriptor: | (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Sweet, M.E, Larsen, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2021-01-09 | | Release date: | 2021-01-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for potassium transport in prokaryotes by KdpFABC.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2BZ2
 
 | | Solution structure of NELF E RRM | | Descriptor: | NEGATIVE ELONGATION FACTOR E | | Authors: | Schweimer, K, Rao, J.N, Neumann, L, Rosch, P, Wohrl, B.M. | | Deposit date: | 2005-08-10 | | Release date: | 2006-08-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies on the RNA-recognition motif of NELF E, a cellular negative transcription elongation factor involved in the regulation of HIV transcription.
Biochem. J., 400, 2006
|
|
1T4R
 
 | | arginase-descarboxy-nor-NOHA complex | | Descriptor: | 3-{[(E)-AMINO(HYDROXYIMINO)METHYL]AMINO}PROPAN-1-AMINIUM, Arginase 1, MANGANESE (II) ION | | Authors: | Cama, E, Pethe, S, Boucher, J.-L, Shoufa, H, Emig, F.A, Ash, D.E, Viola, R.E, Mansuy, D, Christianson, D.W. | | Deposit date: | 2004-04-30 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Inhibitor coordination interactions in the binuclear manganese cluster of arginase
Biochemistry, 43, 2004
|
|
1OON
 
 | | Nitroreductase from e-coli in complex with the dinitrobenzamide prodrug SN27217 | | Descriptor: | 2,4-DINITRO,5-[BIS(2-BROMOETHYL)AMINO]-N-(2',3'-DIOXOPROPYL)BENZAMIDE, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Johansson, E, Parkinson, G.N, Denny, W.A, Neidle, S. | | Deposit date: | 2003-03-04 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Studies on the Nitroreductase Prodrug-Activating System. Crystal Structures of Complexes with the Inhibitor Dicoumarol and Dinitrobenzamide Prodrugs and of the Enzyme Active Form
J.Med.Chem., 46, 2003
|
|
1OO6
 
 | | Nitroreductase from e-coli in complex with the dinitrobenzamide prodrug SN23862 | | Descriptor: | 5-[BIS-2(CHLORO-ETHYL)-AMINO]-2,4-DINTRO-BENZAMIDE, FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Johansson, E, Parkinson, G.N, Denny, W.A, Neidle, S. | | Deposit date: | 2003-03-03 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on the Nitroreductase Prodrug-Activating System. Crystal Structures of Complexes with the Inhibitor Dicoumarol and Dinitrobenzamide Prodrugs and of the Enzyme Active Form
J.Med.Chem., 46, 2003
|
|
3JZ2
 
 | | Crystal structure of human thrombin mutant N143P in E* form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Thrombin heavy chain, ... | | Authors: | Niu, W, Chen, Z, Bush-Pelc, L.A, Bah, A, Gandhi, P.S, Di Cera, E. | | Deposit date: | 2009-09-22 | | Release date: | 2009-10-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mutant N143P reveals how Na+ activates thrombin
J.Biol.Chem., 284, 2009
|
|
1OOQ
 
 | | Nitroreductase from e-coli in complex with the inhibitor dicoumarol | | Descriptor: | BISHYDROXY[2H-1-BENZOPYRAN-2-ONE,1,2-BENZOPYRONE], FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Johansson, E, Parkinson, G.N, Denny, W.A, Neidle, S. | | Deposit date: | 2003-03-04 | | Release date: | 2003-04-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on the Nitroreductase Prodrug-Activating System. Crystal Structures of Complexes with the Inhibitor Dicoumarol and Dinitrobenzamide Prodrugs and of the Enzyme Active Form
J.Med.Chem., 46, 2003
|
|
1T5C
 
 | | Crystal structure of the motor domain of human kinetochore protein CENP-E | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Centromeric protein E, MAGNESIUM ION, ... | | Authors: | Garcia-Saez, I, Yen, T, Wade, R.H, Kozielski, F, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-05-04 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the motor domain of the human kinetochore protein CENP-E.
J.Mol.Biol., 340, 2004
|
|
4Z3P
 
 | | MATE transporter ClbM in complex with Rb+ | | Descriptor: | CACODYLATE ION, Putative drug/sodium antiporter, RUBIDIUM ION | | Authors: | Mousa, J.J, Bruner, S.D. | | Deposit date: | 2015-03-31 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | MATE transport of the E. coli-derived genotoxin colibactin.
Nat Microbiol, 1, 2016
|
|
1IB6
 
 | | CRYSTAL STRUCTURE OF R153C E. COLI MALATE DEHYDROGENASE | | Descriptor: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Bell, J.K, Yennawar, H.P, Wright, S.K, Thompson, J.R, Viola, R.E, Banaszak, L.J. | | Deposit date: | 2001-03-27 | | Release date: | 2001-09-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analyses of a Malate Dehydrogenase with a Variable Active Site
J.Biol.Chem., 276, 2001
|
|
3GOL
 
 | | HCV NS5b polymerase in complex with 1,5 benzodiazepine inhibitor (R)-11d | | Descriptor: | (11R)-10-acetyl-11-(2,4-dichlorophenyl)-6-hydroxy-3,3-dimethyl-2,3,4,5,10,11-hexahydro-1H-dibenzo[b,e][1,4]diazepin-1-one, MAGNESIUM ION, RNA-directed RNA polymerase | | Authors: | Nyanguile, O, De Bondt, H. | | Deposit date: | 2009-03-19 | | Release date: | 2009-06-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | 1,5-Benzodiazepine inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
