8IXV
 
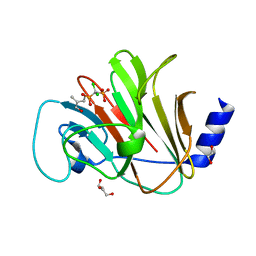 | | Crystal structure of intracellular B30.2 domain of BTN3A in complex with 2Cl-HMBPP | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 3 member A1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, Y.Y, Yi, S.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2023-04-03 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8IZG
 
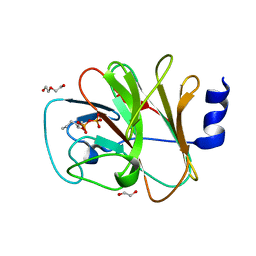 | | Crystal structure of intracellular B30.2 domain of BTN3A1 in complex with 5-HMBPP | | Descriptor: | 1,2-ETHANEDIOL, Butyrophilin subfamily 3 member A1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, Y.Y, Yi, S.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2023-04-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
8JEA
 
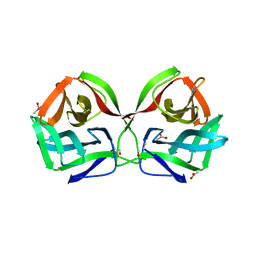 | | Crystal structure of CGL1 from Crassostrea gigas, mannotriose-bound form (CGL1/Man(alpha)1-2Man(alpha)1-2Man) | | Descriptor: | ACETIC ACID, CACODYLATE ION, MAGNESIUM ION, ... | | Authors: | Unno, H, Hatakeyama, T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Mannose oligosaccharide recognition of CGL1, a mannose-specific lectin containing DM9 motifs from Crassostrea gigas, revealed by X-ray crystallographic analysis.
J.Biochem., 175, 2023
|
|
2KA2
 
 | |
8JEB
 
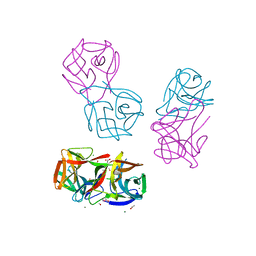 | | Crystal structure of CGL1 from Crassostrea gigas, mannotetraose-bound form (CGL1/Man(alpha)1-2Man(alpha)1-2Man(alpha)1-6Man) | | Descriptor: | ACETIC ACID, MAGNESIUM ION, Natterin-3, ... | | Authors: | Unno, H, Hatakeyama, T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mannose oligosaccharide recognition of CGL1, a mannose-specific lectin containing DM9 motifs from Crassostrea gigas, revealed by X-ray crystallographic analysis.
J.Biochem., 175, 2023
|
|
8JE9
 
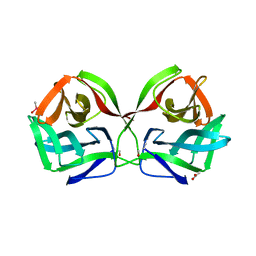 | | Crystal structure of CGL1 from Crassostrea gigas, mannobiose-bound form (CGL1/Man(alpha)1-2Man) | | Descriptor: | ACETIC ACID, CACODYLATE ION, Natterin-3, ... | | Authors: | Unno, H, Hatakeyama, T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Mannose oligosaccharide recognition of CGL1, a mannose-specific lectin containing DM9 motifs from Crassostrea gigas, revealed by X-ray crystallographic analysis.
J.Biochem., 175, 2023
|
|
8JDA
 
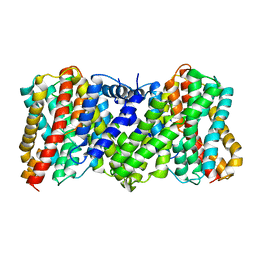 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class2 | | Descriptor: | Sodium/hydrogen exchanger 7 | | Authors: | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
8JD9
 
 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Sodium/hydrogen exchanger 7 | | Authors: | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
8J1V
 
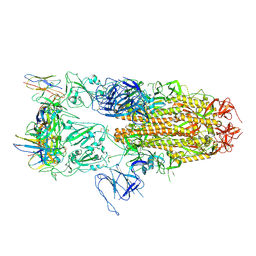 | |
2KCN
 
 | | Solution structure of the antifungal protein PAF from Penicillium chrysogenum | | Descriptor: | Antifungal protein | | Authors: | Batta, G, Barna, T, Gaspari, Z, Sandor, S, Kover, K.E, Binder, U, Sarg, B, Kaiserer, L, Chhillar, A.K, Eigentler, A, Leiter, E, Hegedus, N, Pocsi, I, Lindner, H, Marx, F. | | Deposit date: | 2008-12-23 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Functional aspects of the solution structure and dynamics of PAF--a highly-stable antifungal protein from Penicillium chrysogenum
Febs J., 276, 2009
|
|
7OR6
 
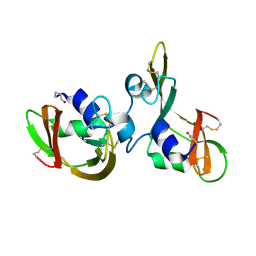 | |
7ORD
 
 | |
7P2Q
 
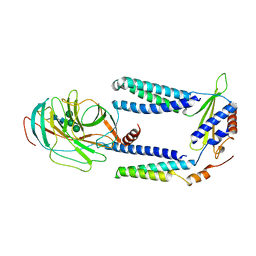 | | Human Signal Peptidase Complex Paralog C (SPC-C) | | Descriptor: | Signal peptidase complex catalytic subunit SEC11C, Signal peptidase complex subunit 1, Signal peptidase complex subunit 2, ... | | Authors: | Liaci, A.M, Foerster, F. | | Deposit date: | 2021-07-06 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure of the human signal peptidase complex reveals the determinants for signal peptide cleavage.
Mol.Cell, 81, 2021
|
|
7P2P
 
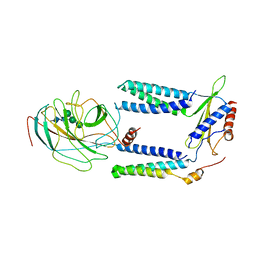 | | Human Signal Peptidase Complex Paralog A (SPC-A) | | Descriptor: | Signal peptidase complex catalytic subunit SEC11A, Signal peptidase complex subunit 1, Signal peptidase complex subunit 2, ... | | Authors: | Liaci, A.M, Foerster, F. | | Deposit date: | 2021-07-06 | | Release date: | 2021-10-06 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure of the human signal peptidase complex reveals the determinants for signal peptide cleavage.
Mol.Cell, 81, 2021
|
|
8J54
 
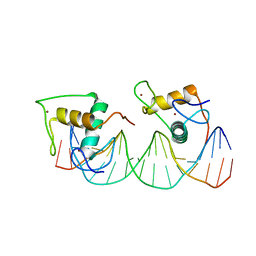 | | Crystal structure of RXR/DR2 complex | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*CP*CP*TP*AP*CP*TP*GP*AP*CP*CP*TP*AP*G)-3'), DNA (5'-D(*CP*TP*AP*GP*GP*TP*CP*AP*GP*TP*AP*GP*GP*TP*CP*AP*TP*G)-3'), Retinoic acid receptor RXR, ... | | Authors: | Chen, Y, Jiang, L. | | Deposit date: | 2023-04-21 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural characterization of the DNA binding mechanism of retinoic acid-related orphan receptor gamma.
Structure, 32, 2024
|
|
7OWY
 
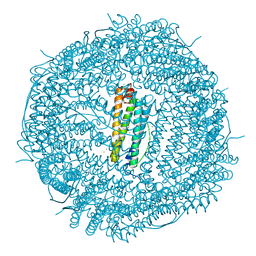 | | Crystal structure of human mitochondrial ferritin (hMTF) Fe(II)-loaded for 3 minutes showing a peroxide anion as bridging species of iron ions in the ferroxidase site | | Descriptor: | CHLORIDE ION, FE (II) ION, Ferritin, ... | | Authors: | Pozzi, C, Ciambellotti, S, Tassone, G, Turano, P, Mangani, S. | | Deposit date: | 2021-06-21 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Iron Binding in the Ferroxidase Site of Human Mitochondrial Ferritin.
Chemistry, 27, 2021
|
|
7P06
 
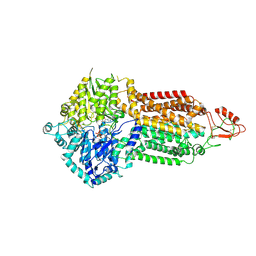 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in outward-facing conformation with ADP-orthovanadate/ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADP ORTHOVANADATE, MAGNESIUM ION, ... | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P05
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation with ADP/ATP and rhodamine 6G | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Pleiotropic ABC efflux transporter of multiple drugs, ... | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
8J9G
 
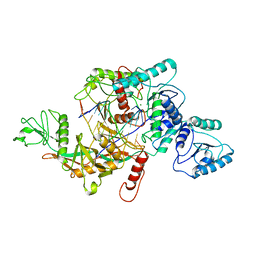 | | CrtSPARTA hetero-dimer bound with guide-target, state 1 | | Descriptor: | DNA (25-MER), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Li, Z.X, Guo, L.J, Huang, P.P, Xiao, Y.B, Chen, M.R. | | Deposit date: | 2023-05-03 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system.
Nat.Chem.Biol., 20, 2024
|
|
7P03
 
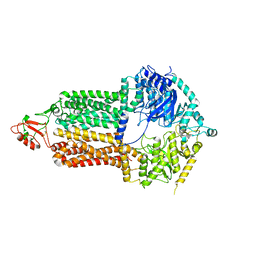 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation without nucleotides | | Descriptor: | Pleiotropic ABC efflux transporter of multiple drugs | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P04
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation with ADP/ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Pleiotropic ABC efflux transporter of multiple drugs | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
8J8H
 
 | | SPARTA monomer bound with guide-target, state 2 | | Descriptor: | DNA (25-MER), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Li, Z.X, Guo, L.J, Huang, P.P, Xiao, Y.B, Chen, M.R. | | Deposit date: | 2023-05-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system.
Nat.Chem.Biol., 20, 2024
|
|
7P47
 
 | | Structure of the E3 ligase Smc5/Nse2 in complex with Ubc9-SUMO thioester mimetic | | Descriptor: | E3 SUMO-protein ligase MMS21, SUMO-conjugating enzyme UBC9, Structural maintenance of chromosomes protein 5, ... | | Authors: | Lascorz, J, Varejao, N, Reverter, D. | | Deposit date: | 2021-07-09 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.314 Å) | | Cite: | Structural basis for the E3 ligase activity enhancement of yeast Nse2 by SUMO-interacting motifs.
Nat Commun, 12, 2021
|
|
8J9P
 
 | | SPARTA dimer bound with guide-target | | Descriptor: | DNA (25-MER), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Li, Z.X, Guo, L.J, Huang, P.P, Xiao, Y.B, Chen, M.R. | | Deposit date: | 2023-05-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Auto-inhibition and activation of a short Argonaute-associated TIR-APAZ defense system.
Nat.Chem.Biol., 20, 2024
|
|
8J9F
 
 | | Structure of STG-hydrolyzing beta-glucosidase 1 (PSTG1) | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Yanai, T, Imaizumi, R, Takahashi, Y, Katsumura, E, Yamamoto, M, Nakayama, T, Yamashita, S, Takeshita, K, Sakai, N, Matsuura, H. | | Deposit date: | 2023-05-03 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into a bacterial beta-glucosidase capable of degrading sesaminol triglucoside to produce sesaminol: toward the understanding of the aglycone recognition mechanism by the C-terminal lid domain.
J.Biochem., 174, 2023
|
|
