7OM4
 
 | |
7OY6
 
 | | Crystal structure of human DYRK1A in complex with ARN25068 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, ~{N}4-(3-cyclopropyl-1~{H}-pyrazol-5-yl)-~{N}2-(phenylmethyl)thieno[3,2-d]pyrimidine-2,4-diamine | | Authors: | Tripathi, S.K, Balboni, B, Demuro, S, DiMartino, R, Ortega, J, Girotto, S, Cavalli, A. | | Deposit date: | 2021-06-23 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | ARN25068, a versatile starting point towards triple GSK-3 beta /FYN/DYRK1A inhibitors to tackle tau-related neurological disorders.
Eur.J.Med.Chem., 229, 2022
|
|
1AIZ
 
 | | STRUCTURE OF APO-AZURIN FROM ALCALIGENES DENITRIFICANS AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, CADMIUM ION, SULFATE ION | | Authors: | Baker, E.N, Anderson, B.F, Blackwell, K.A. | | Deposit date: | 1993-11-11 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of apo-azurin from Alcaligenes denitrificans at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
7ADP
 
 | | Cryo-EM structure of human ER membrane protein complex in GDN detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ER membrane protein complex subunit 1, ER membrane protein complex subunit 10, ... | | Authors: | Braeuning, B, Prabu, J.R, Miller-Vedam, L.E, Weissman, J.S, Frost, A, Schulman, B.A. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Elife, 9, 2020
|
|
6Q8P
 
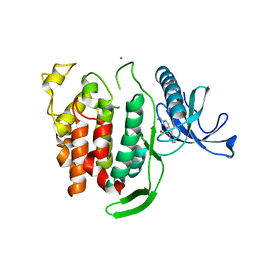 | | Structure of CLK1 with bound N-methyl-10-nitropyrido[3,4-g]quinazolin-2-amine | | Descriptor: | Dual specificity protein kinase CLK1, POTASSIUM ION, ~{N}-methyl-10-nitro-pyrido[3,4-g]quinazolin-2-amine | | Authors: | Joerger, A.C, Chatterjee, D, Schroeder, M, Tazarki, H, Zeinyeh, W, Esvan, Y.J, Khiari, J, Josselin, B, Baratte, B, Bach, S, Ruchaud, S, Anizon, F, Giraud, F, Moreau, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New pyrido[3,4-g]quinazoline derivatives as CLK1 and DYRK1A inhibitors: synthesis, biological evaluation and binding mode analysis.
Eur J Med Chem, 166, 2019
|
|
6Q11
 
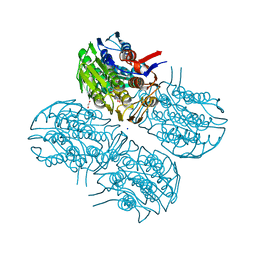 | | Crystal structure of MurA from Clostridium difficile, mutation C116S, in the presence of URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Dopkins, B.J, Call, C.J, Thoden, J.B, Holden, H.M. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of MurA from Clostridium difficile, mutation C116S, in the presence of URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID
To Be Published
|
|
6XG8
 
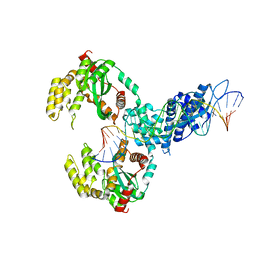 | | ISCth4 transposase, pre-cleaved complex, PCC | | Descriptor: | DNA (26-MER), Mutator family transposase | | Authors: | Kosek, D, Dyda, F. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of ISCth4 transpososomes reveal the role of asymmetry in copy-out/paste-in DNA transposition.
Embo J., 40, 2021
|
|
8GPK
 
 | |
5JHW
 
 | |
8GJU
 
 | | Crystal structure of human methylmalonyl-CoA mutase (MMUT) in complex with methylmalonic acidemia type A protein (MMAA), coenzyme A, and GDP | | Descriptor: | COENZYME A, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Mascarenhas, R.M, Ruetz, M, Gouda, H, Yaw, M, Banerjee, R. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Architecture of the human G-protein-methylmalonyl-CoA mutase nanoassembly for B 12 delivery and repair.
Nat Commun, 14, 2023
|
|
6JO6
 
 | | Structure of the green algal photosystem I supercomplex with light-harvesting complex I | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Miyazaki, N, Takahashi, Y. | | Deposit date: | 2019-03-20 | | Release date: | 2019-06-19 | | Last modified: | 2019-06-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the green algal photosystem I supercomplex with a decameric light-harvesting complex I.
Nat.Plants, 5, 2019
|
|
6Q8K
 
 | | CLK1 with bound pyridoquinazoline | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase CLK1, ~{N}2-(3-morpholin-4-ylpropyl)pyrido[3,4-g]quinazoline-2,10-diamine | | Authors: | Schroeder, M, Tazarki, H, Zeinyeh, W, Esvan, Y.J, Khiari, J, Joesselin, B, Bach, S, Ruchaud, S, Anizon, F, Giraud, F, Moreau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | New pyrido[3,4-g]quinazoline derivatives as CLK1 and DYRK1A inhibitors: synthesis, biological evaluation and binding mode analysis.
Eur J Med Chem, 166, 2019
|
|
7NZA
 
 | | Structure of OBP1 from Varroa destructor, form P2<1> | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Odorant Binding Protein from Varroa destructor, form P2<1>, ... | | Authors: | Cambillau, C, Amigues, B, Roussel, A, Leone, P, Gaubert, A, Pelosi, P. | | Deposit date: | 2021-03-23 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | A new non-classical fold of varroa odorant-binding proteins reveals a wide open internal cavity.
Sci Rep, 11, 2021
|
|
6VLI
 
 | | Crystal structure of transcriptional regulator from bacteriophage 186 | | Descriptor: | ACETATE ION, ACETIC ACID, Regulatory protein CII, ... | | Authors: | Truong, J.Q, Panjikar, S, Bruning, J.B, Shearwin, K.E. | | Deposit date: | 2020-01-24 | | Release date: | 2021-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.100021 Å) | | Cite: | Crystal structure of a transcriptional regulator from bacteriophage 186
To Be Published
|
|
7U5O
 
 | | CRYSTAL STRUCTURE OF THE BONE MORPHOGENETIC PROTEIN RECEPTOR TYPE 2 LIGAND BINDING DOMAIN IN COMPLEX WITH ACTIVIN-B | | Descriptor: | Bone morphogenetic protein receptor type-2, Inhibin beta B chain | | Authors: | Chu, K.Y, Malik, A, Thamilselvan, V, Martinez-Hackert, E. | | Deposit date: | 2022-03-02 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Type II BMP and activin receptors BMPR2 and ACVR2A share a conserved mode of growth factor recognition.
J.Biol.Chem., 298, 2022
|
|
6VTX
 
 | | Crystal structure of human KLF4 zinc finger DNA binding domain in complex with NANOG DNA | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*GP*GP*TP*GP*TP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*AP*CP*AP*CP*CP*CP*CP*CP*T)-3'), Krueppel-like factor 4, ... | | Authors: | Sharma, R, Sharma, S, Choi, K.J, Ferreon, A.C.M, Ferreon, J.C, Sankaran, B, MacKenzie, K.R, Kim, C. | | Deposit date: | 2020-02-13 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Liquid condensation of reprogramming factor KLF4 with DNA provides a mechanism for chromatin organization.
Nat Commun, 12, 2021
|
|
7U92
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML1006a | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-03-09 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
6PTM
 
 | |
5K5E
 
 | | Discovery and Structure-Activity Relationships of a Highly Selective Butyrylcholinesterase Inhibitor by Structure-Based Virtual Screening | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | De la Mora, E, Dighe, S.N, Deora, G.S, Ross, B.P, Nachon, F, Brazzolotto, X. | | Deposit date: | 2016-05-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Structure-Activity Relationships of a Highly Selective Butyrylcholinesterase Inhibitor by Structure-Based Virtual Screening.
J.Med.Chem., 59, 2016
|
|
6Q41
 
 | | Atomic resolution crystal structure of a BAA collagen heterotrimer | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Leading chain of the BAA collagen heterotrimer, ... | | Authors: | Jalan, A.A, Hartgerink, J.D, Brear, P, Leitinger, B, Farndale, R.W. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Atomic resolution crystal structure of an AAB collagen heterotrimer
Nat.Chem.Biol., 2019
|
|
7OFQ
 
 | |
4YFF
 
 | | TNNI3K complexed with inhibitor 2 | | Descriptor: | 3-[(5-bromo-7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]-N-methyl-4-(morpholin-4-yl)benzenesulfonamide, Serine/threonine-protein kinase TNNI3K | | Authors: | Shewchuk, L.M, Wang, L, Lawhorn, B.G. | | Deposit date: | 2015-02-25 | | Release date: | 2015-09-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Identification of Purines and 7-Deazapurines as Potent and Selective Type I Inhibitors of Troponin I-Interacting Kinase (TNNI3K).
J.Med.Chem., 58, 2015
|
|
6NIH
 
 | | Crystal structure of human TLR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Su, L, Zhang, H. | | Deposit date: | 2018-12-27 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of TLR2/TLR1 Activation by the Synthetic Agonist Diprovocim.
J. Med. Chem., 62, 2019
|
|
6T26
 
 | | X-ray crystal structure of Vibrio alkaline phosphatase with the non-competitive inhibitor cyclohexylamine | | Descriptor: | Alkaline phosphatase, CHLORIDE ION, CYCLOHEXYLAMMONIUM ION, ... | | Authors: | Asgeirsson, B, Hjorleifsson, J.G, Markusson, S, Helland, R. | | Deposit date: | 2019-10-07 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.265 Å) | | Cite: | X-ray crystal structure of Vibrio alkaline phosphatase with the non-competitive inhibitor cyclohexylamine.
Biochem Biophys Rep, 24, 2020
|
|
6ZI8
 
 | | X-ray diffraction structure of bovine insulin at 2.3 A resolution | | Descriptor: | CHLORIDE ION, Insulin, ZINC ION | | Authors: | Housset, D, Ling, W.L, Bacia-Verloop, M, Zander, U, McCarthy, A.A, Schoehn, G. | | Deposit date: | 2020-06-25 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
