5SYJ
 
 | |
3IQD
 
 | | Structure of Octopine-dehydrogenase in complex with NADH and Agmatine | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, AGMATINE, Octopine dehydrogenase | | Authors: | Smits, S.H.J, Meyer, T, Mueller, A, Willbold, D, Grieshaber, M.K, Schmitt, L. | | Deposit date: | 2009-08-20 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into the mechanism of ligand binding to octopine dehydrogenase from Pecten maximus by NMR and crystallography
Plos One, 5, 2010
|
|
5V6P
 
 | | CryoEM structure of the ERAD-associated E3 ubiquitin-protein ligase HRD1 | | Descriptor: | ERAD-associated E3 ubiquitin-protein ligase HRD1 | | Authors: | Schoebel, S, Mi, W, Stein, A, Rapoport, T.A, Liao, M. | | Deposit date: | 2017-03-17 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3.
Nature, 548, 2017
|
|
7JL4
 
 | | Crystal structure of TRIM65 PSpry domain | | Descriptor: | GLYCEROL, Tripartite motif-containing protein 65 | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
2FER
 
 | | P450CAM from Pseudomonas putida reconstituted with manganic protoporphyrin IX | | Descriptor: | Cytochrome P450-cam, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING MN | | Authors: | von Koenig, K, Makris, T.M, Sligar, S.G, Schlichting, I. | | Deposit date: | 2005-12-16 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The status of high-valent metal oxo complexes in the P450 cytochromes.
J.Inorg.Biochem., 100, 2006
|
|
7JL2
 
 | | Cryo-EM structure of MDA5-dsRNA filament in complex with TRIM65 PSpry domain (Trimer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
8U45
 
 | | Crystal Structure Analysis of Aspergillus fumigatus alkaline protease | | Descriptor: | Alkaline protease 1, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fernandez, D, Diec, D.D.L, Guo, W, Russi, S. | | Deposit date: | 2023-09-08 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting Aspergillus allergen oryzin with a chemical probe at atomic precision.
Sci Rep, 13, 2023
|
|
7JL0
 
 | | Cryo-EM structure of MDA5-dsRNA in complex with TRIM65 PSpry domain (Monomer) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | Authors: | Kato, K, Ahmad, S, Hur, S. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases.
Mol.Cell, 81, 2021
|
|
6VK0
 
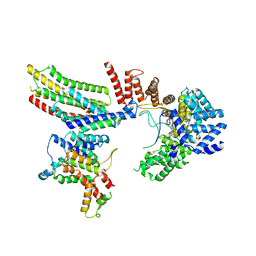 | | CryoEM structure of Hrd1-Usa1/Der1/Hrd3 of the flipped topology | | Descriptor: | Degradation in the endoplasmic reticulum protein 1, ERAD-associated E3 ubiquitin-protein ligase HRD1, ERAD-associated E3 ubiquitin-protein ligase component HRD3, ... | | Authors: | Wu, X, Rapoport, T.A. | | Deposit date: | 2020-01-18 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex.
Science, 368, 2020
|
|
2BKM
 
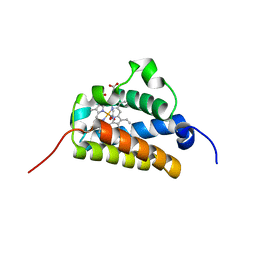 | | Crystal structure of the truncated hemoglobin from Geobacillus stearothermophilus | | Descriptor: | ACETATE ION, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ilari, A, Kjelgaard, P, von Wachenfeldt, C, Boffi, A, Chiancone, E. | | Deposit date: | 2006-02-08 | | Release date: | 2006-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure and Ligand Binding Properties of the Truncated Hemoglobin from Geobacillus Stearothermophilus
Arch.Biochem.Biophys., 457, 2007
|
|
3BY8
 
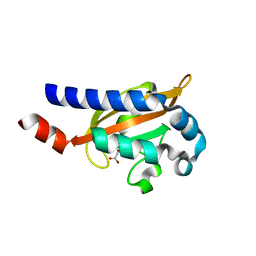 | | Crystal Structure of the E.coli DcuS Sensor Domain | | Descriptor: | (2S)-2-hydroxybutanedioic acid, Sensor protein dcuS | | Authors: | Cheung, J, Hendrickson, W.A. | | Deposit date: | 2008-01-15 | | Release date: | 2008-08-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of C4-Dicarboxylate Ligand Complexes with Sensor Domains of Histidine Kinases DcuS and DctB.
J.Biol.Chem., 283, 2008
|
|
5G1B
 
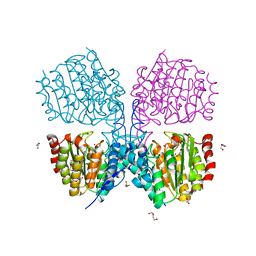 | | Bordetella Alcaligenes HDAH native | | Descriptor: | DI(HYDROXYETHYL)ETHER, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, PENTAETHYLENE GLYCOL, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-24 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The thermodynamic signature of ligand binding to histone deacetylase-like amidohydrolases is most sensitive to the flexibility in the L2-loop lining the active site pocket.
Biochim. Biophys. Acta, 1861, 2017
|
|
8QN2
 
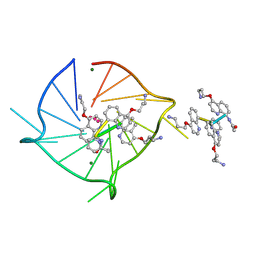 | |
5TJG
 
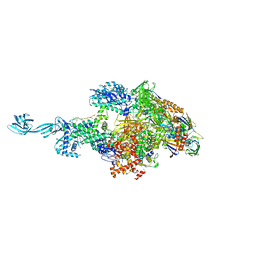 | |
2YZ3
 
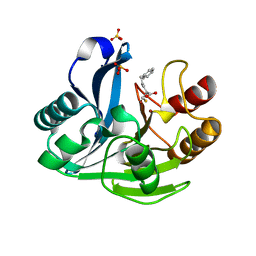 | | Crystallographic Investigation of Inhibition Mode of the VIM-2 Metallo-beta-lactamase from Pseudomonas aeruginosa with Mercaptocarboxylate Inhibitor | | Descriptor: | (S)-2-(MERCAPTOMETHYL)-5-PHENYLPENTANOIC ACID, Metallo-beta-lactamase, SULFATE ION, ... | | Authors: | Yamaguchi, Y, Yamagata, Y, Arakawa, Y, Kurosaki, H. | | Deposit date: | 2007-05-02 | | Release date: | 2008-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic investigation of the inhibition mode of a VIM-2 metallo-beta-lactamase from Pseudomonas aeruginosa by a mercaptocarboxylate inhibitor.
J.Med.Chem., 50, 2007
|
|
5E6W
 
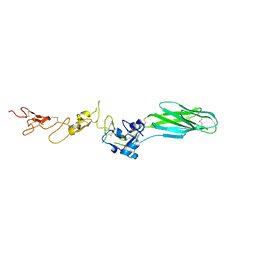 | |
5SYI
 
 | | Structure of D141A variant of B. pseudomallei KatG complexed with INH | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Catalase-peroxidase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2016-08-11 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of the antitubercular pro-drug isoniazid in the heme access channel of catalase-peroxidase (KatG). A combined structural and metadynamics investigation.
J Phys Chem B, 118, 2014
|
|
5G17
 
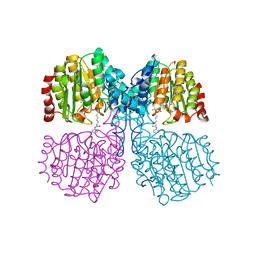 | | Bordetella Alcaligenes HDAH (T101A) bound to 9,9,9-trifluoro-8,8- dihydroxy-N-phenylnonanamide. | | Descriptor: | 9,9,9-tris(fluoranyl)-8,8-bis(oxidanyl)-~{N}-phenyl-nonanamide, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, POTASSIUM ION, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-23 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The thermodynamic signature of ligand binding to histone deacetylase-like amidohydrolases is most sensitive to the flexibility in the L2-loop lining the active site pocket.
Biochim. Biophys. Acta, 1861, 2017
|
|
7WN2
 
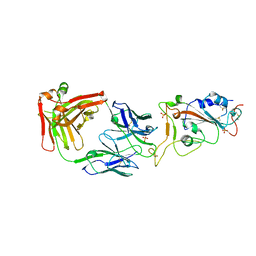 | |
1XJ7
 
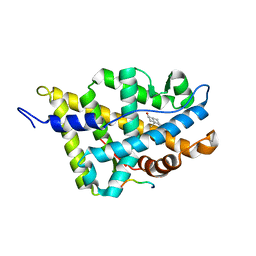 | | Complex Androgen Receptor LBD and RAC3 peptide | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, RAC3 derived peptide | | Authors: | Estebanez-Perpina, E, Moore, J.M.R, Mar, E, Nguyen, P, Delgado-Rodrigues, E, Baxter, J.D, Buehrer, B.M, Webb, P, Fletterick, R.J, Guy, R.K. | | Deposit date: | 2004-09-22 | | Release date: | 2005-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Molecular Mechanisms of Coactivator Utilization in Ligand-dependent Transactivation by the Androgen Receptor
J.Biol.Chem., 280, 2005
|
|
2BGV
 
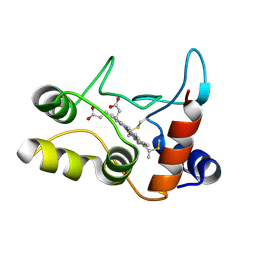 | | X-ray structure of ferric cytochrome c-550 from Paracoccus versutus | | Descriptor: | CYTOCHROME C-550, HEME C | | Authors: | Worrall, J.A.R, Van Roon, A.-M.M, Ubbink, M, Canters, G.W. | | Deposit date: | 2005-01-05 | | Release date: | 2005-05-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Effect of Replacing the Axial Methionine Ligand with a Lysine Residue in Cytochrome C-550 from Paracoccus Versutus Assessed by X-Ray Crystallography and Unfolding.
FEBS J., 272, 2005
|
|
4J2T
 
 | | Inhibitor-bound Ca2+ ATPase | | Descriptor: | (3S,3aR,4S,6S,6aR,7S,8S,9bS)-6-(acetyloxy)-3a,4-bis(butanoyloxy)-3-hydroxy-3,6,9-trimethyl-8-{[(2E)-2-methylbut-2-enoyl]oxy}-2-oxo-2,3,3a,4,5,6,6a,7,8,9b-decahydroazuleno[4,5-b]furan-7-yl octanoate, PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, ... | | Authors: | Paulsen, E.S, Villadsen, J, Tenori, E, Liu, H, Lie, M.A, Bonde, D.F, Bublitz, M, Olesen, C, Autzen, H.E, Dach, I, Sehgal, P, Moller, J.V, Schiott, B, Nissen, P, Christensen, S.B. | | Deposit date: | 2013-02-05 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Water-mediated interactions influence the binding of thapsigargin to sarco/endoplasmic reticulum calcium adenosinetriphosphatase.
J.Med.Chem., 56, 2013
|
|
3BRI
 
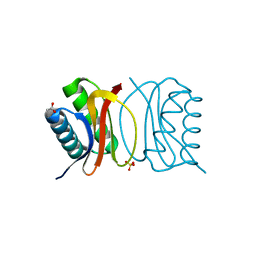 | | Crystal Structure of apo-LC8 | | Descriptor: | ACETATE ION, Dynein light chain 1, cytoplasmic, ... | | Authors: | Benison, G, Karplus, P.A, Barbar, E, Chiodo, M. | | Deposit date: | 2007-12-21 | | Release date: | 2008-12-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Interplay of Ligand Binding and Quaternary Structure in the Diverse Interactions of Dynein Light Chain LC8.
J.Mol.Biol., 384, 2008
|
|
3BY9
 
 | |
7WNB
 
 | |
