8BWS
 
 | | Structure of yeast RNA Polymerase III elongation complex at 3.3 A | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Nguyen, P.Q, Fernandez-Tornero, C. | | Deposit date: | 2022-12-07 | | Release date: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nat Commun, 14, 2023
|
|
6DZX
 
 | | Crystal structure of the N. meningitides methionine-binding protein in its D-methionine bound conformation. | | Descriptor: | D-METHIONINE, Lipoprotein | | Authors: | Nguyen, P.T, Lai, J.Y, Kaiser, J.T, Rees, D.C. | | Deposit date: | 2018-07-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.678 Å) | | Cite: | Structures of the Neisseria meningitides methionine-binding protein MetQ in substrate-free form and bound to l- and d-methionine isomers.
Protein Sci., 28, 2019
|
|
7Z2Z
 
 | | Structure of yeast RNA Polymerase III-DNA-Ty1 integrase complex (Pol III-DNA-IN1) at 3.1 A | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Nguyen, P.Q, Fernandez-Tornero, C. | | Deposit date: | 2022-03-01 | | Release date: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nat Commun, 14, 2023
|
|
6CVA
 
 | |
8D3X
 
 | | Human alpha3 Na+/K+-ATPase in its K+-occluded state | | Descriptor: | FXYD domain-containing ion transport regulator 6, Sodium/potassium-transporting ATPase subunit alpha-3, Sodium/potassium-transporting ATPase subunit beta-1, ... | | Authors: | Nguyen, P.T, Bai, X. | | Deposit date: | 2022-06-01 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for gating mechanism of the human sodium-potassium pump.
Nat Commun, 13, 2022
|
|
7Z0H
 
 | | Structure of yeast RNA Polymerase III-Ty1 integrase complex at 2.6 A (focus subunit AC40). | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Nguyen, P.Q, Huecas, S, Plaza-Pegueroles, A, Fernandez-Tornero, C. | | Deposit date: | 2022-02-22 | | Release date: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nat Commun, 14, 2023
|
|
7Z30
 
 | |
7Z31
 
 | | Structure of yeast RNA Polymerase III-Ty1 integrase complex at 2.7 A (focus subunit C11, no C11 C-terminal Zn-ribbon in the funnel pore). | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Nguyen, P.Q, Huecas, S, Plaza-Pegueroles, A, Fernandez-Tornero, C. | | Deposit date: | 2022-03-01 | | Release date: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nat Commun, 14, 2023
|
|
8D3U
 
 | | Human alpha3 Na+/K+-ATPase in its Na+-occluded state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FXYD domain-containing ion transport regulator 6, Sodium/potassium-transporting ATPase subunit alpha-3, ... | | Authors: | Nguyen, P.T, Bai, X. | | Deposit date: | 2022-06-01 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for gating mechanism of the human sodium-potassium pump.
Nat Commun, 13, 2022
|
|
8D3Y
 
 | | Human alpha3 Na+/K+-ATPase in its exoplasmic side-open state | | Descriptor: | FXYD domain-containing ion transport regulator 6, Sodium/potassium-transporting ATPase subunit alpha-3, Sodium/potassium-transporting ATPase subunit beta-1 | | Authors: | Nguyen, P.T, Bai, X. | | Deposit date: | 2022-06-01 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for gating mechanism of the human sodium-potassium pump.
Nat Commun, 13, 2022
|
|
8D3W
 
 | |
8D3V
 
 | | Human alpha3 Na+/K+-ATPase in its cytoplasmic side-open state | | Descriptor: | FXYD domain-containing ion transport regulator 6, Sodium/potassium-transporting ATPase subunit alpha-3, Sodium/potassium-transporting ATPase subunit beta-1 | | Authors: | Nguyen, P.T, Bai, X. | | Deposit date: | 2022-06-01 | | Release date: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for gating mechanism of the human sodium-potassium pump.
Nat Commun, 13, 2022
|
|
6OJA
 
 | | Crystal structure of the N. meningitides methionine-binding protein in its L-methionine bound conformation | | Descriptor: | Lipoprotein, METHIONINE | | Authors: | Nguyen, P.T, Lai, J.Y, Kaiser, J.T, Rees, D.C. | | Deposit date: | 2019-04-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of the Neisseria meningitides methionine-binding protein MetQ in substrate-free form and bound to l- and d-methionine isomers.
Protein Sci., 28, 2019
|
|
6CVL
 
 | | Crystal structure of the Escherichia coli ATPgS-bound MetNI methionine ABC transporter in complex with its MetQ binding protein | | Descriptor: | IODIDE ION, MERCURY (II) ION, MetI transmembrane subunit, ... | | Authors: | Nguyen, P.T, Kaiser, J.T, Rees, D.C. | | Deposit date: | 2018-03-28 | | Release date: | 2018-11-14 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.953 Å) | | Cite: | Noncanonical role for the binding protein in substrate uptake by the MetNI methionine ATP Binding Cassette (ABC) transporter.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5OCJ
 
 | | Crystal structure of Ag85C bound to cyclophostin 8beta inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, Diacylglycerol acyltransferase/mycolyltransferase Ag85C, methoxy-[(3~{R})-3-[(2~{R})-1-methoxy-1,3-bis(oxidanylidene)butan-2-yl]pentadecyl]phosphinic acid | | Authors: | Viljoen, A, Richard, M, Nguyen, P.C, Spilling, C.D, Canaan, S, Cavalier, J.F, Blaise, M, Kremer, L. | | Deposit date: | 2017-07-03 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclipostins and cyclophostin analogs inhibit the antigen 85C from
J. Biol. Chem., 293, 2018
|
|
1T65
 
 | | Crystal structure of the androgen receptor ligand binding domain with DHT and a peptide derived form its physiological coactivator GRIP1 NR box 2 bound in a non-helical conformation | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 2 | | Authors: | Estebanez-Perpina, E, Moore, J.M.R, Mar, E, Nguyen, P, Delgado-Rodrigues, E, Baxter, J.D, Webb, P, Fletterick, R.J, Guy, R.K. | | Deposit date: | 2004-05-05 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The Molecular Mechanisms of Coactivator Utilization in Ligand-dependent Transactivation by the Androgen Receptor.
J.Biol.Chem., 280, 2005
|
|
1T5Z
 
 | | Crystal Structure of the Androgen Receptor Ligand Binding Domain (LBD) with DHT and a peptide derived from its physiological coactivator ARA70 | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 4 | | Authors: | Estebanez-Perpina, E, Moore, J.M.R, Mar, E, Nguyen, P, Delgado-Rodrigues, E, Baxter, J.D, Webb, P, Fletterick, R.J, Guy, R.K. | | Deposit date: | 2004-05-05 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Molecular Mechanisms of Coactivator Utilization in Ligand-dependent Transactivation by the Androgen Receptor.
J.Biol.Chem., 280, 2005
|
|
1T63
 
 | | Crystal Structure of the Androgen Receptor Ligand Binding Domain with DHT and a peptide derived from its physiological coactivator GRIP1 NR box3 | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 2 | | Authors: | Estebanez-Perpina, E, Moore, J.M.R, Mar, E, Nguyen, P, Delgado-Rodrigues, E, Baxter, J.D, Webb, P, Fletterick, R.J, Guy, R.K. | | Deposit date: | 2004-05-05 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The Molecular Mechanisms of Coactivator Utilization in Ligand-dependent Transactivation by the Androgen Receptor.
J.Biol.Chem., 280, 2005
|
|
1XJ7
 
 | | Complex Androgen Receptor LBD and RAC3 peptide | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, RAC3 derived peptide | | Authors: | Estebanez-Perpina, E, Moore, J.M.R, Mar, E, Nguyen, P, Delgado-Rodrigues, E, Baxter, J.D, Buehrer, B.M, Webb, P, Fletterick, R.J, Guy, R.K. | | Deposit date: | 2004-09-22 | | Release date: | 2005-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Molecular Mechanisms of Coactivator Utilization in Ligand-dependent Transactivation by the Androgen Receptor
J.Biol.Chem., 280, 2005
|
|
2N5Q
 
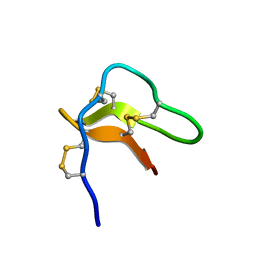 | | Solution structure of cystein-rich peptide jS1 from Jasminum sambac | | Descriptor: | cysteine-rich peptide jS1 | | Authors: | Shin, J, Kumari, G, Serra, A, Nguyen, P.Q.T, Yoon, H, Tam, J.P. | | Deposit date: | 2015-07-24 | | Release date: | 2016-07-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification of a cysteine-rich peptide family with unusual disulfide connectivity from Jasminum sambac
To be Published
|
|
4BFH
 
 | |
3TUZ
 
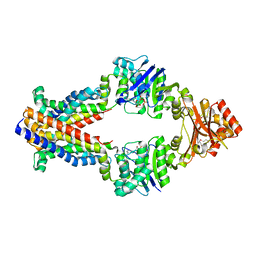 | | Inward facing conformations of the MetNI methionine ABC transporter: CY5 SeMet soak crystal form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-methionine transport system permease protein metI, Methionine import ATP-binding protein MetN, ... | | Authors: | Johnson, E, Nguyen, P, Rees, D.C. | | Deposit date: | 2011-09-19 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Inward facing conformations of the MetNI methionine ABC transporter: Implications for the mechanism of transinhibition.
Protein Sci., 21, 2012
|
|
3TUJ
 
 | |
3TUI
 
 | |
3CPZ
 
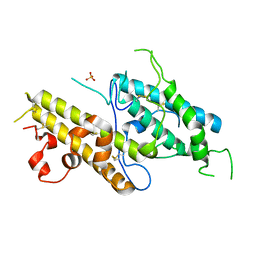 | | Crystal structure of VAR2CSA DBL3x domain in the presence of dodecasaccharide of CSA | | Descriptor: | Erythrocyte membrane protein 1, SULFATE ION | | Authors: | Singh, K, Gittis, A.G, Nguyen, P, Gowda, D.C, Miller, L.H, Garboczi, D.N. | | Deposit date: | 2008-04-01 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the DBL3x domain of pregnancy-associated malaria protein VAR2CSA complexed with chondroitin sulfate A.
Nat.Struct.Mol.Biol., 15, 2008
|
|
