1S9G
 
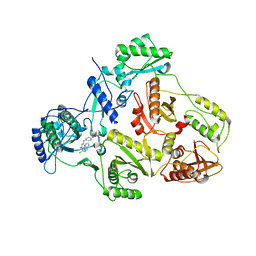 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R120394. | | Descriptor: | 4-[4-AMINO-6-(5-CHLORO-1H-INDOL-4-YLMETHYL)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
5JU4
 
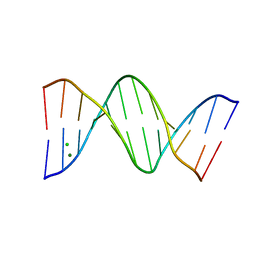 | |
3ECO
 
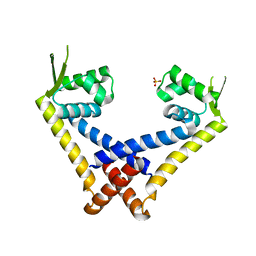 | |
1K2Z
 
 | |
1JTL
 
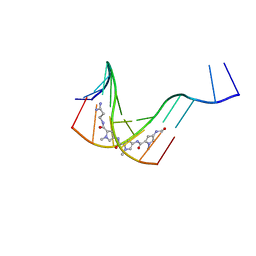 | |
3GJH
 
 | | Crystal structure of a DNA duplex containing 7,8-dihydropyridol[2,3-d]pyrimidin-2-one | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(B7C)P*GP*CP*G)-3', 6-AMIDINE-2-(4-AMIDINO-PHENYL)INDOLE | | Authors: | Takenaka, A, Juan, E.C.M, Shimizu, S, Haraguchi, T, Xiao, M, Kurose, T. | | Deposit date: | 2009-03-09 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into the stabilizing contributions of bicyclic cytosine analogues: crystal structures of DNA duplexes containing 7,8-dihydropyridol[2,3-d]pyrimidin-2-one
To be Published
|
|
8GU7
 
 | |
8GT9
 
 | |
3AJJ
 
 | | Crystal Structure of d(CGCGGATf5UCGCG): 5-Formyluridine/Guanosine Base-pair in B-DNA | | Descriptor: | 5'-D(*CP*GP*CP*GP*GP*AP*TP*(UFR)P*CP*GP*CP*G)-3' | | Authors: | Tsunoda, M, Sakaue, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2010-06-07 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Insights into the structures of DNA damaged by hydroxyl radical: crystal structures of DNA duplexes containing 5-formyluracil
J Nucleic Acids, 2010, 2010
|
|
3AJK
 
 | | Crystal structure of d(CGCGGATf5UCGCG): 5-Formyluridine:Guanosine Base-pair in B-DNA with Hoechst33258 | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, 5'-D(*CP*GP*CP*GP*GP*AP*TP*(UFR)P*CP*GP*CP*G*)-3', MAGNESIUM ION | | Authors: | Tsunoda, M, Sakaue, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2010-06-07 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the structures of DNA damaged by hydroxyl radical: crystal structures of DNA duplexes containing 5-formyluracil
J Nucleic Acids, 2010, 2010
|
|
4G1Q
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with Rilpivirine (TMC278, Edurant), a non-nucleoside rt-inhibiting drug | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, MAGNESIUM ION, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2012-07-11 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Snapshot of the equilibrium dynamics of a drug bound to HIV-1 reverse transcriptase.
Nat Chem, 5, 2013
|
|
7WIY
 
 | | Cryo-EM structure of human TPH2 tetramer | | Descriptor: | FE (III) ION, IMIDAZOLE, Tryptophan 5-hydroxylase 2 | | Authors: | Zhu, K.F, Liu, C, Zhang, H.W, Wang, D.P. | | Deposit date: | 2022-01-05 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryo-EM Structure and Activator Screening of Human Tryptophan Hydroxylase 2.
Front Pharmacol, 13, 2022
|
|
8P2D
 
 | | Cryo-EM structure of the dimeric form of the anaerobic ribonucleotide reductase from Prevotella copri produced in the presence of dATP and CTP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, MAGNESIUM ION | | Authors: | Banerjee, I, Bimai, O, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-09-13 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Nucleotide binding to the ATP-cone in anaerobic ribonucleotide reductases allosterically regulates activity by modulating substrate binding.
Elife, 12, 2024
|
|
5MUJ
 
 | | BT0996 RGII Chain B Complex | | Descriptor: | ACETATE ION, Beta-galactosidase, alpha-L-rhamnopyranose-(1-2)-[alpha-L-rhamnopyranose-(1-3)]alpha-L-arabinopyranose-(1-4)-[4-O-[(1R)-1-hydroxyethyl]-2-O-methyl-alpha-L-fucopyranose-(1-2)]beta-D-galactopyranose-(1-2)-alpha-D-aceric acid-(1-4)-alpha-L-rhamnopyranose-(1-3)-3-C-(hydroxylmethyl)-alpha-D-erythrofuranose | | Authors: | Cartmell, A, Basle, A, Ndeh, D, Luis, A.S, Venditto, I, Labourel, A, Rogowski, A, Gilbert, H.J. | | Deposit date: | 2017-01-13 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
3QF2
 
 | | Crystal structure of NALP3 PYD | | Descriptor: | NACHT, LRR and PYD domains-containing protein 3 | | Authors: | Park, H.H, Bae, J.Y. | | Deposit date: | 2011-01-21 | | Release date: | 2011-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of NALP3 Protein Pyrin Domain (PYD) and Its Implications in Inflammasome Assembly
J.Biol.Chem., 286, 2011
|
|
3MKN
 
 | | Crystal structure of the E. coli pyrimidine nucleosidase YeiK bound to a competitive inhibitor | | Descriptor: | (2S,3S,4R,5R)-2-(3,4-diaminophenyl)-5-(hydroxymethyl)pyrrolidine-3,4-diol, CALCIUM ION, Putative uncharacterized protein YeiK | | Authors: | Garau, G, Fornili, A, Giabbai, B, Degano, M. | | Deposit date: | 2010-04-15 | | Release date: | 2010-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Energy Landscapes Associated with Macromolecular Conformational Changes from Endpoint Structures
J.Am.Chem.Soc., 132, 2010
|
|
2MPC
 
 | |
5IBQ
 
 | | Crystal structure of an ABC solute binding protein from Rhizobium etli CFN 42 (RHE_PF00037,TARGET EFI-511357) in complex with alpha-D-apiose | | Descriptor: | 3-C-(hydroxylmethyl)-alpha-D-erythrofuranose, CALCIUM ION, Probable ribose ABC transporter, ... | | Authors: | Vetting, M.W, Carter, M.S, Al Obaidi, N.F, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of an ABC solute binding protein from Rhizobium etli CFN 42 (RHE_PF00037,TARGET EFI-511357) in complex with alpha-D-apiose
To be published
|
|
2NN6
 
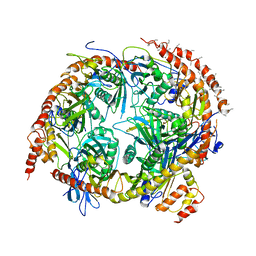 | | Structure of the human RNA exosome composed of Rrp41, Rrp45, Rrp46, Rrp43, Mtr3, Rrp42, Csl4, Rrp4, and Rrp40 | | Descriptor: | 3'-5' exoribonuclease CSL4 homolog, Exosome complex exonuclease RRP4, Exosome complex exonuclease RRP40, ... | | Authors: | Lima, C.D. | | Deposit date: | 2006-10-23 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Reconstitution, activities, and structure of the eukaryotic RNA exosome.
Cell(Cambridge,Mass.), 127, 2006
|
|
4KS8
 
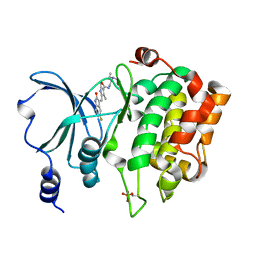 | | PAK6 kinase domain in complex with sunitinib | | Descriptor: | N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide, Serine/threonine-protein kinase PAK 6 | | Authors: | Gao, J, Boggon, T.J. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate and Inhibitor Specificity of the Type II p21-Activated Kinase, PAK6.
Plos One, 8, 2013
|
|
4KS7
 
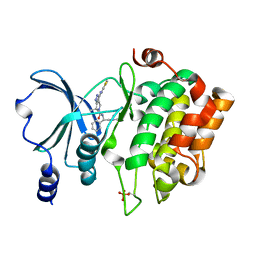 | | PAK6 kinase domain in complex with PF-3758309 | | Descriptor: | ISOPROPYL ALCOHOL, PF-3758309, Serine/threonine-protein kinase PAK 6 | | Authors: | Gao, J, Boggon, T.J. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Substrate and Inhibitor Specificity of the Type II p21-Activated Kinase, PAK6.
Plos One, 8, 2013
|
|
8HWQ
 
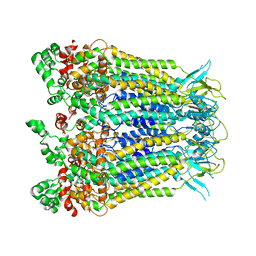 | |
8K53
 
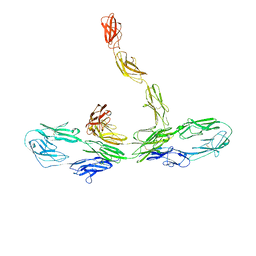 | |
8K3J
 
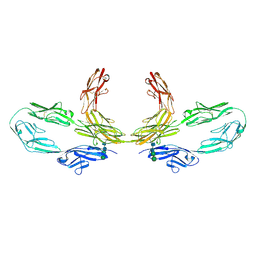 | | Structure of human CNTN2 immunoglobulin domains 1-6 homo-dimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-2, ... | | Authors: | Zhang, Z.Z. | | Deposit date: | 2023-07-16 | | Release date: | 2024-07-17 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human CNTN2 immunoglobulin domains 1-6 homo-dimer
To Be Published
|
|
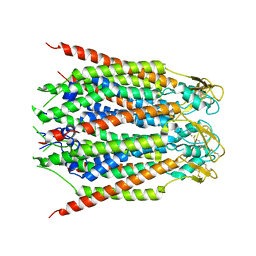
8GN8
 
 | |
