3MCV
 
 | | Structure of PTR1 from Trypanosoma brucei in ternary complex with 2,4-diamino-5-[2-(2,5-dimethoxyphenyl)ethyl]thieno[2,3-d]-pyrimidine and NADP+ | | Descriptor: | 5-[2-(2,5-dimethoxyphenyl)ethyl]thieno[2,3-d]pyrimidine-2,4-diamine, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Tulloch, L.B, Hunter, W.N. | | Deposit date: | 2010-03-29 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-resolution structures of Trypanosoma brucei pteridine reductase ligand complexes inform on the placement of new molecular entities in the active site of a potential drug target.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3MUT
 
 | | Crystal Structure of the G20A/C92U mutant c-di-GMP riboswith bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), G20A/C92U mutant c-di-GMP riboswitch, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
2BAW
 
 | | Human Nuclear Receptor-Ligand Complex 1 | | Descriptor: | Peroxisome proliferator activated receptor delta, VACCENIC ACID, heptyl beta-D-glucopyranoside | | Authors: | Fyffe, S.A, Alphey, M.S, Buetow, L, Smith, T.K, Ferguson, M.A.J, Sorensen, M.D, Bjorkling, F, Hunter, W.N. | | Deposit date: | 2005-10-15 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reevaluation of the PPAR-beta/delta Ligand Binding Domain Model Reveals Why It Exhibits the Activated Form
Mol.Cell, 21, 2006
|
|
6M69
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with GMPPCP (GDP) | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, Hydrolase, ... | | Authors: | Raj, P, Karthik, S, Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Plasticity, ligand conformation and enzyme action of Mycobacterium smegmatis MutT1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
3MUR
 
 | | Crystal Structure of the C92U mutant c-di-GMP riboswith bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), C92U mutant c-di-GMP riboswitch, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
6JP5
 
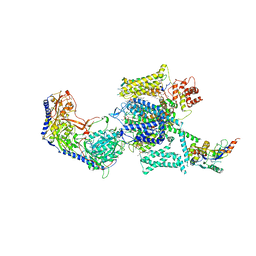 | | Rabbit Cav1.1-Nifedipine Complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Huang, G, Wu, J, Yan, N. | | Deposit date: | 2019-03-25 | | Release date: | 2019-06-12 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular Basis for Ligand Modulation of a Mammalian Voltage-Gated Ca2+Channel.
Cell, 177, 2019
|
|
6FKP
 
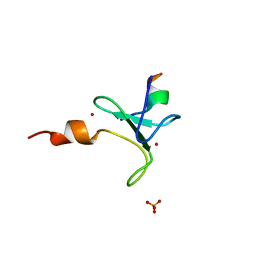 | | Crystal structure of BAZ2A PHD zinc finger in complex with H3 10-mer AA mutant peptide | | Descriptor: | ALA-ARG-THR-ALA-ALA-THR-ALA-ARG, Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, ... | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2018-01-24 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
1SEJ
 
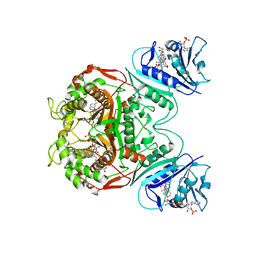 | | Crystal Structure of Dihydrofolate Reductase-Thymidylate Synthase from Cryptosporidium hominis Bound to 1843U89/NADPH/dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, S)-2-(5(((1,2-DIHYDRO-3-METHYL-1-OXOBENZO(F)QUINAZOLIN-9-YL)METHYL)AMINO)1-OXO-2-ISOINDOLINYL)GLUTARIC ACID, ... | | Authors: | Anderson, A.C. | | Deposit date: | 2004-02-17 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Two crystal structures of dihydrofolate reductase-thymidylate synthase from Cryptosporidium hominis reveal protein-ligand interactions including a structural basis for observed antifolate resistance.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
8U83
 
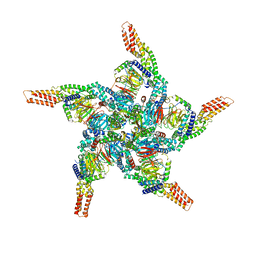 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State C From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.975 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U84
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State D From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3MUM
 
 | | Crystal Structure of the G20A mutant c-di-GMP riboswith bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), G20A mutant c-di-GMP Riboswitch, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
3MUV
 
 | | Crystal Structure of the G20A/C92U mutant c-di-GMP riboswith bound to c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, G20A/C92U mutant c-di-GMP riboswitch, MAGNESIUM ION, ... | | Authors: | Strobel, S.A, Smith, K.D. | | Deposit date: | 2010-05-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch .
Biochemistry, 49, 2010
|
|
2H28
 
 | | Crystal structure of YeeU from E. coli. Northeast Structural Genomics target ER304 | | Descriptor: | CHLORIDE ION, GLYCEROL, Hypothetical protein yeeU, ... | | Authors: | Arbing, M, Su, M, Benach, J, Karpowich, N.K, Jiang, M, Xiao, R, Cunningham, K, Ma, L.-C, Chen, C.X, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-18 | | Release date: | 2006-07-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
5PZK
 
 | |
5PZL
 
 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE IN COMPLEX WITH 2-({3-[1-(2-CYCLOPROPYLETHYL)-6-FLUORO-4-HYDROXY-2-OXO-1,2-DIHYDROQUINOLIN-3-YL]-1,1-DIOXO-1,4-DIHYDRO-1LAMBDA~6~,2,4-BENZOTHIADIAZIN-7-YL}OXY)ACETAMIDE | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, 2-({3-[1-(2-cyclopropylethyl)-6-fluoro-4-hydroxy-2-oxo-1,2-dihydroquinolin-3-yl]-1,1-dioxo-1,4-dihydro-1lambda~6~,2,4-benzothiadiazin-7-yl}oxy)acetamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-02-27 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Discovery of a Hepatitis C Virus NS5B Replicase Palm Site Allosteric Inhibitor (BMS-929075) Advanced to Phase 1 Clinical Studies.
J. Med. Chem., 60, 2017
|
|
4RPG
 
 | |
6FHQ
 
 | | Crystal structure of human BAZ2B PHD zinc finger in complex with Fr 21 | | Descriptor: | 2-azanyl-~{N}-(1,3-thiazol-2-yl)ethanamide, Bromodomain adjacent to zinc finger domain protein 2B, ZINC ION | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2018-01-15 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
5B5G
 
 | | Crystal structure of ALiS4-Streptavidin complex | | Descriptor: | SULFITE ION, Streptavidin, methyl 5-(4-oxidanylidene-5~{H}-furo[3,2-c]pyridin-2-yl)pyridine-3-carboxylate | | Authors: | Sugiyama, S, Terai, T, Kakinouchi, K, Fujikake, R, Nagano, T, Urano, Y. | | Deposit date: | 2016-05-04 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Improving the Solubility of Artificial Ligands of Streptavidin to Enable More Practical Reversible Switching of Protein Localization in Cells
Chembiochem, 18, 2017
|
|
6FHU
 
 | | Crystal structure of BAZ2A PHD zinc finger in complex with H3 3-mer peptide | | Descriptor: | ALA-ARG-TAM, Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, ... | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2018-01-15 | | Release date: | 2018-03-21 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
8ZRN
 
 | | Structure of abt | | Descriptor: | 5-[[(2~{S})-azetidin-2-yl]methoxy]-2-chloranyl-pyridine, Neuronal acetylcholine receptor subunit alpha-6, Neuronal acetylcholine receptor subunit beta-4,Soluble cytochrome b562, ... | | Authors: | Su, J, Yu, Z, Zhao, Y. | | Deposit date: | 2024-06-04 | | Release date: | 2025-04-30 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Molecular insights into the alpha 6 beta 4 nicotinic acetylcholine receptor function and ligand recognition.
Nat Commun, 16, 2025
|
|
8ZRP
 
 | | Structure of nico | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, Neuronal acetylcholine receptor subunit alpha-6, Neuronal acetylcholine receptor subunit beta-4,Soluble cytochrome b562, ... | | Authors: | Su, J, Yu, Z, Zhao, Y. | | Deposit date: | 2024-06-05 | | Release date: | 2025-04-30 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Molecular insights into the alpha 6 beta 4 nicotinic acetylcholine receptor function and ligand recognition.
Nat Commun, 16, 2025
|
|
4RPK
 
 | | Crystal structure of Micobacterium tuberculosis UDP-Galactopyranose mutase in complex with tetrafluorinated substrate analog UDP-F4-Galf | | Descriptor: | (2R,5S)-5-[(1R)-1,2-dihydroxyethyl]-3,3,4,4-tetrafluorotetrahydrofuran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name), FLAVIN-ADENINE DINUCLEOTIDE, UDP-galactopyranose mutase | | Authors: | Van Straaten, K.E, Sanders, D.A.R. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis of Ligand Binding to UDP-Galactopyranose Mutase from Mycobacterium tuberculosis Using Substrate and Tetrafluorinated Substrate Analogues.
J.Am.Chem.Soc., 137, 2015
|
|
4RPH
 
 | |
6FI1
 
 | | Crystal structure of human BAZ2B PHD zinc finger in complex with Fr23 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, ZINC ION, ~{N}-(4-aminophenyl)-2-azanyl-ethanamide | | Authors: | Amato, A, Lucas, X, Bortoluzzi, A, Wright, D, Ciulli, A. | | Deposit date: | 2018-01-16 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting Ligandable Pockets on Plant Homeodomain (PHD) Zinc Finger Domains by a Fragment-Based Approach.
ACS Chem. Biol., 13, 2018
|
|
4RPL
 
 | | Crystal structure of Micobacterium tuberculosis UDP-Galactopyranose mutase in complex with tetrafluorinated substrate analog UDP-F4-Galp | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, UDP-galactopyranose mutase, [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,5S,6R)-3,3,4,4-tetrafluoro-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl dihydrogen diphosphate (non-preferred name) | | Authors: | Van Straaten, K.E, Sanders, D.A.R. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2499 Å) | | Cite: | Structural Basis of Ligand Binding to UDP-Galactopyranose Mutase from Mycobacterium tuberculosis Using Substrate and Tetrafluorinated Substrate Analogues.
J.Am.Chem.Soc., 137, 2015
|
|
