4VUB
 
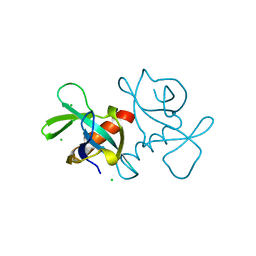 | | CCDB, A TOPOISOMERASE POISON FROM ESCHERICHIA COLI | | Descriptor: | CCDB, CHLORIDE ION | | Authors: | Loris, R, Dao-Thi, M.-H, Bahasi, E.M, Van Melderen, L, Poortmans, F, Liddington, R, Couturier, M, Wyns, L. | | Deposit date: | 1998-04-17 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of CcdB, a topoisomerase poison from E. coli.
J.Mol.Biol., 285, 1999
|
|
6NIP
 
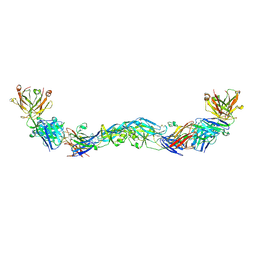 | | Crystal structure of a human anti-ZIKV-DENV neutralizing antibody MZ1 in complex with ZIKV E glycoprotein | | Descriptor: | Envelope protein E, MZ1 Heavy chain, MZ1 Light Chain | | Authors: | Sankhala, R.S, Dussupt, V, Donofrio, G, Choe, M, Modjarrad, K, Michael, N.L, Krebs, S.J, Joyce, M.G. | | Deposit date: | 2018-12-31 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4.16 Å) | | Cite: | Potent Zika and dengue cross-neutralizing antibodies induced by Zika vaccination in a dengue-experienced donor.
Nat Med, 26, 2020
|
|
4V8U
 
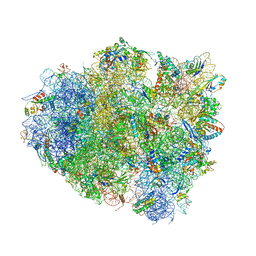 | | Crystal Structure of 70S Ribosome with Both Cognate tRNAs in the E and P Sites Representing an Authentic Elongation Complex. | | Descriptor: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, Y.G, Feng, S, Chen, Y. | | Deposit date: | 2012-08-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of 70S ribosome with both cognate tRNAs in the E and P sites representing an authentic elongation complex.
PLoS ONE, 8, 2013
|
|
6ND5
 
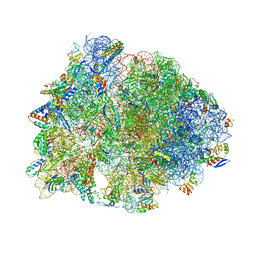 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with chloramphenicol and bound to mRNA and A-, P-, and E-site tRNAs at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Plessa, E, Chen, C.-W, Bougas, A, Krokidis, M.G, Dinos, G.P, Polikanov, Y.S. | | Deposit date: | 2018-12-13 | | Release date: | 2019-03-20 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High-resolution crystal structures of ribosome-bound chloramphenicol and erythromycin provide the ultimate basis for their competition.
RNA, 25, 2019
|
|
6ND6
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with erythromycin and bound to mRNA and A-, P-, and E-site tRNAs at 2.85A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Plessa, E, Chen, C.-W, Bougas, A, Krokidis, M.G, Dinos, G.P, Polikanov, Y.S. | | Deposit date: | 2018-12-13 | | Release date: | 2019-02-20 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | High-resolution crystal structures of ribosome-bound chloramphenicol and erythromycin provide the ultimate basis for their competition.
RNA, 25, 2019
|
|
4V97
 
 | | Crystal structure of the bacterial ribosome ram mutation G299A. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fagan, C.E, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2012-04-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.516 Å) | | Cite: | Reorganization of an intersubunit bridge induced by disparate 16S ribosomal ambiguity mutations mimics an EF-Tu-bound state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4V7J
 
 | | Structure of RelE nuclease bound to the 70S ribosome (precleavage state) | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Neubauer, C, Gao, Y.-G, Andersen, K.R, Dunham, C.M, Kelley, A.C, Hentschel, J, Gerdes, K, Ramakrishnan, V, Brodersen, D.E. | | Deposit date: | 2009-11-02 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE.
Cell(Cambridge,Mass.), 139, 2009
|
|
5W4K
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with Klebsazolicin and bound to mRNA and A-, P- and E-site tRNAs at 2.7A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Metelev, M, Osterman, I.A, Ghilarov, D, Khabibullina, N.F, Yakimov, A, Shabalin, K, Utkina, I, Travin, D.Y, Komarova, E.S, Serebryakova, M, Artamonova, T, Khodorkovskii, M, Konevega, A.L, Sergiev, P.V, Severinov, K, Polikanov, Y.S. | | Deposit date: | 2017-06-12 | | Release date: | 2017-08-30 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Klebsazolicin inhibits 70S ribosome by obstructing the peptide exit tunnel.
Nat. Chem. Biol., 13, 2017
|
|
8IWH
 
 | | Structure and characteristics of a photosystem II supercomplex containing monomeric LHCX and dimeric FCPII antennae from the diatom Thalassiosira pseudonana | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15-octaen-17-ynyl]cyclohex-3-en-1-ol, (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, ... | | Authors: | Feng, Y, Li, Z.H, Wang, W.D, Shen, J.R. | | Deposit date: | 2023-03-30 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structure of a diatom photosystem II supercomplex containing a member of Lhcx family and dimeric FCPII.
Sci Adv, 9, 2023
|
|
4V4N
 
 | | Structure of the Methanococcus jannaschii ribosome-SecYEBeta channel complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein L7AE, ... | | Authors: | Menetret, J.F, Park, E, Gumbart, J.C, Ludtke, S.J, Li, W, Whynot, A, Rapoport, T.A, Akey, C.W. | | Deposit date: | 2013-06-17 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure of the SecY channel during initiation of protein translocation.
Nature, 506, 2013
|
|
5LS7
 
 | | Complex of wild type E. coli alpha aspartate decarboxylase with its processing factor PanZ | | Descriptor: | ACETYL COENZYME *A, Aspartate 1-decarboxylase, CARBON DIOXIDE, ... | | Authors: | Monteiro, D.C.F, Webb, M.E, Pearson, A.R. | | Deposit date: | 2016-08-22 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | The Mechanism of Regulation of Pantothenate Biosynthesis by the PanD-PanZAcCoA Complex Reveals an Additional Mode of Action for the Antimetabolite N-Pentyl Pantothenamide (N5-Pan).
Biochemistry, 56, 2017
|
|
6ZZY
 
 | | Structure of high-light grown Chlorella ohadii photosystem I | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ... | | Authors: | Caspy, I, Nelson, N, Nechushtai, R, Shkolnisky, Y, Neumann, E. | | Deposit date: | 2020-08-05 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
7A4P
 
 | | Structure of small high-light grown Chlorella ohadii photosystem I | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (3R)-beta,beta-caroten-3-ol, ... | | Authors: | Caspy, I, Nelson, N, Nechushtai, R, Shkolnisky, Y, Neumann, E. | | Deposit date: | 2020-08-20 | | Release date: | 2021-07-28 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
5LXS
 
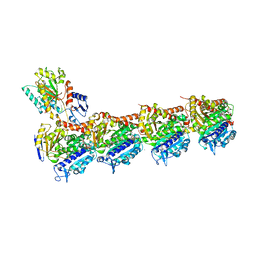 | | Tubulin-KS-1-199-32 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Steinmetz, M.O. | | Deposit date: | 2016-09-22 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Microtubule Stabilization by Discodermolide.
Chembiochem, 18, 2017
|
|
6ZZX
 
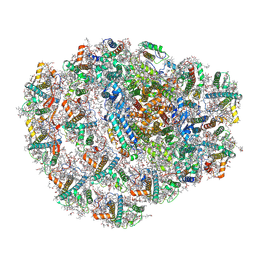 | | Structure of low-light grown Chlorella ohadii Photosystem I | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, (3R)-beta,beta-caroten-3-ol, ... | | Authors: | Caspy, I, Nelson, N, Nechushtai, R, Neumann, E, Shkolnisky, Y. | | Deposit date: | 2020-08-05 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM photosystem I structure reveals adaptation mechanisms to extreme high light in Chlorella ohadii.
Nat.Plants, 7, 2021
|
|
6NFZ
 
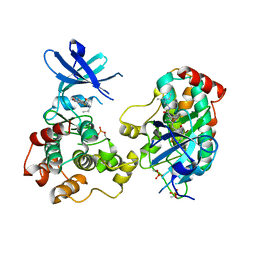 | | Crystal structure of diphosphorylated HPK1 kinase domain in complex with sunitinib in the active state. | | Descriptor: | Mitogen-activated protein kinase kinase kinase kinase 1, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide | | Authors: | Johnson, E, McTigue, M, Cronin, C.N. | | Deposit date: | 2018-12-21 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.966 Å) | | Cite: | Multiple conformational states of the HPK1 kinase domain in complex with sunitinib reveal the structural changes accompanying HPK1 trans-regulation.
J.Biol.Chem., 294, 2019
|
|
6ZQM
 
 | | bovine ATP synthase monomer state 2 (combined) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-07-10 | | Release date: | 2020-09-09 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5LI3
 
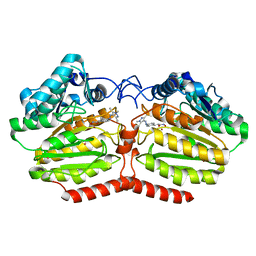 | | Crystal structure of HDAC-like protein from P. aeruginosa in complex with a photo-switchable inhibitor. | | Descriptor: | (2E)-N-hydroxy-3-{4-[(E)-(1,3,5-trimethyl-1H-pyrazol-4-yl)diazenyl]phenyl}prop-2-enamide, Acetoin utilization protein, POTASSIUM ION, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-07-14 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Toward Photopharmacological Antimicrobial Chemotherapy Using Photoswitchable Amidohydrolase Inhibitors.
ACS Infect Dis, 3, 2017
|
|
5LNT
 
 | | Crystal structure of Arabidopsis thaliana Pdx1K166R-preI320 complex | | Descriptor: | PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.1, [(~{E},4~{S})-4-azanyl-3-oxidanylidene-pent-1-enyl] dihydrogen phosphate | | Authors: | Rodrigues, M.J, Windeisen, V, Zhang, Y, Guedez, G, Weber, S, Strohmeier, M, Hanes, J.W, Royant, A, Evans, G, Sinning, I, Ealick, S.E, Begley, T.P, Tews, I. | | Deposit date: | 2016-08-06 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Lysine relay mechanism coordinates intermediate transfer in vitamin B6 biosynthesis.
Nat. Chem. Biol., 13, 2017
|
|
7Y7B
 
 | | Cryo-EM structure of cryptophyte photosystem I | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, ... | | Authors: | Zhao, L.S, Li, K, Zhang, Y.Z, Liu, L.N. | | Deposit date: | 2022-06-22 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis and evolution of the photosystem I-light-harvesting supercomplex of cryptophyte algae.
Plant Cell, 35, 2023
|
|
1LWC
 
 | | CRYSTAL STRUCTURE OF M184V MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE, PHOSPHATE ION | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
7Y8A
 
 | | Cryo-EM structure of cryptophyte photosystem I | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, ... | | Authors: | Zhao, L.S, Zhang, Y.Z, Liu, L.N, Li, K. | | Deposit date: | 2022-06-23 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural basis and evolution of the photosystem I-light-harvesting supercomplex of cryptophyte algae.
Plant Cell, 35, 2023
|
|
7YCA
 
 | | Cryo-EM structure of the PSI-LHCI-Lhcp supercomplex from Ostreococcus tauri | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (1~{S})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaenyl]cyclohex-3-en-1-ol, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Shan, J, Sheng, X, Ishii, A, Watanabe, A, Song, C, Murata, K, Minagawa, J, Liu, Z. | | Deposit date: | 2022-07-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | The photosystem I supercomplex from a primordial green alga Ostreococcus tauri harbors three light-harvesting complex trimers.
Elife, 12, 2023
|
|
1LWF
 
 | | CRYSTAL STRUCTURE OF A MUTANT HIV-1 REVERSE TRANSCRIPTASE (RTMQ+M184V: M41L/D67N/K70R/M184V/T215Y) IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
4G1Q
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with Rilpivirine (TMC278, Edurant), a non-nucleoside rt-inhibiting drug | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, MAGNESIUM ION, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2012-07-11 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Snapshot of the equilibrium dynamics of a drug bound to HIV-1 reverse transcriptase.
Nat Chem, 5, 2013
|
|
