8WA3
 
 | | Cryo-EM structure of peptide free and Gs-coupled GIPR | | Descriptor: | Gastric inhibitory polypeptide receptor,Fusion protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1,O-antigen polymerase, ... | | Authors: | Cong, Z.T, Zhao, F.H, Li, Y, Luo, G, Zhou, Q.T, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-09-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Molecular features of the ligand-free GLP-1R, GCGR and GIPR in complex with G s proteins.
Cell Discov, 10, 2024
|
|
8EYQ
 
 | | 30S_delta_ksgA_h44_inactive_conformation | | Descriptor: | 16S_rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Ortega, J, Sun, J. | | Deposit date: | 2022-10-28 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | KsgA facilitates ribosomal small subunit maturation by proofreading a key structural lesion.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8WG8
 
 | | Cryo-EM structures of peptide free and Gs-coupled GCGR | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cong, Z.T, Zhao, F.H, Li, Y, Luo, G, Zhou, Q.T, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-09-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Molecular features of the ligand-free GLP-1R, GCGR and GIPR in complex with G s proteins.
Cell Discov, 10, 2024
|
|
8WG7
 
 | | Cryo-EM structures of peptide free and Gs-coupled GLP-1R | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Cong, Z.T, Zhao, F.H, Li, Y, Luo, G, Zhou, Q.T, Yang, D.H, Wang, M.W. | | Deposit date: | 2023-09-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Molecular features of the ligand-free GLP-1R, GCGR and GIPR in complex with G s proteins.
Cell Discov, 10, 2024
|
|
8EFW
 
 | |
8EFX
 
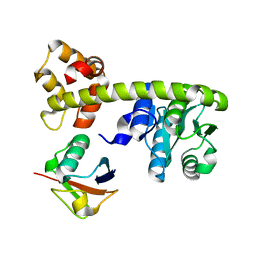 | |
8EYT
 
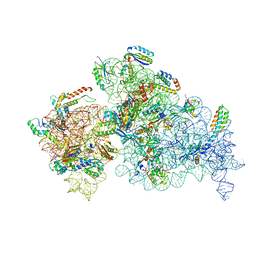 | | 30S_delta_ksgA+KsgA complex | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Sun, J, Kinman, L.F, Jahagirdar, D, Ortega, J, Davis, J.H. | | Deposit date: | 2022-10-28 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | KsgA facilitates ribosomal small subunit maturation by proofreading a key structural lesion.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8PIV
 
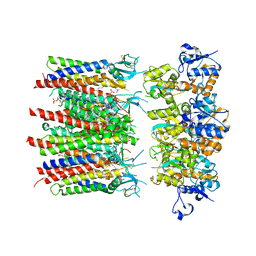 | | Homomeric GluA2 flip R/G-unedited Q/R-edited F231A mutant in tandem with TARP gamma-2, desensitized conformation 1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Glutamate receptor, ... | | Authors: | Zhang, D, Krieger, J.M, Yamashita, K, Greger, I.H. | | Deposit date: | 2023-06-22 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor.
Nature, 621, 2023
|
|
1B4O
 
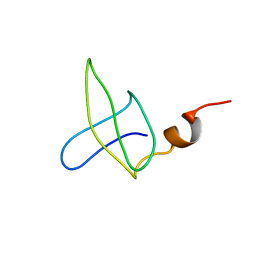 | |
8PYQ
 
 | | 10 micrometer HEWL crystals solved at room-temperature using fixed-target serial crystallography. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Mason, T.J, Carrillo, M, Beale, J.H, Padeste, C. | | Deposit date: | 2023-07-25 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Micro-structured polymer fixed targets for serial crystallography at synchrotrons and XFELs.
Iucrj, 10, 2023
|
|
8PYO
 
 | | 5 micrometer HEWL crystals solved at room-temperature using fixed-target serial crystallography. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Mason, T.J, Carrillo, M, Beale, J.H, Padeste, C. | | Deposit date: | 2023-07-25 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Micro-structured polymer fixed targets for serial crystallography at synchrotrons and XFELs.
Iucrj, 10, 2023
|
|
8PYP
 
 | | 25 micrometer HEWL crystals solved at room-temperature using fixed-target serial crystallography. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Mason, T.J, Carrillo, M, Beale, J.H, Padeste, C. | | Deposit date: | 2023-07-25 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Micro-structured polymer fixed targets for serial crystallography at synchrotrons and XFELs.
Iucrj, 10, 2023
|
|
5ZCW
 
 | | Structure of the Methanosarcina mazei class II CPD-photolyase in complex with intact, phosphodiester linked, CPD-lesion | | Descriptor: | 5'-D(*AP*TP*CP*GP*GP*CP*(TTD)P*CP*GP*CP*GP*CP*AP*A)-3', 5'-D(*TP*GP*CP*GP*CP*GP*AP*AP*GP*CP*CP*GP*AP*T)-3', ACETATE ION, ... | | Authors: | Maestre-Reyna, M, Bessho, Y. | | Deposit date: | 2018-02-21 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Twist and turn: a revised structural view on the unpaired bubble of class II CPD photolyase in complex with damaged DNA.
IUCrJ, 5, 2018
|
|
8W9A
 
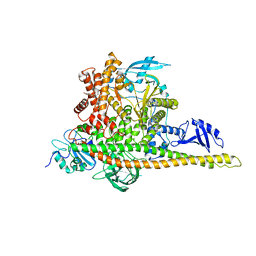 | | CryoEM structure of human PI3K-alpha (P85/P110-H1047R) with QR-7909 binding at an allosteric site | | Descriptor: | 6-chloranyl-3-[[(1R)-1-[2-(1,3-dihydropyrrolo[3,4-c]pyridin-2-yl)-3,6-dimethyl-4-oxidanylidene-quinazolin-8-yl]ethyl]amino]pyridine-2-carboxylic acid, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Huang, X, Ren, X, Zhong, W. | | Deposit date: | 2023-09-05 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures reveal two allosteric inhibition modes of PI3K alpha H1047R involving a re-shaping of the activation loop.
Structure, 32, 2024
|
|
1BHL
 
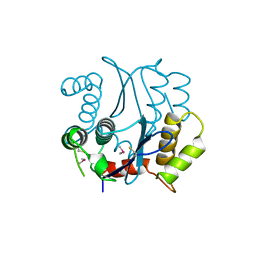 | | CACODYLATED CATALYTIC DOMAIN OF HIV-1 INTEGRASE | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Maignan, S, Guilloteau, J.P, Zhou-Liu, Q, Clement-Mella, C, Mikol, V. | | Deposit date: | 1998-06-10 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases.
J.Mol.Biol., 282, 1998
|
|
1AQQ
 
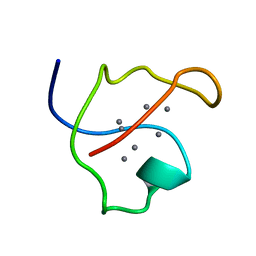 | | AG-SUBSTITUTED METALLOTHIONEIN FROM SACCHAROMYCES CEREVISIAE, NMR, 10 STRUCTURES | | Descriptor: | AG-METALLOTHIONEIN, SILVER ION | | Authors: | Peterson, C.W, Narula, S.S, Armitage, I.M. | | Deposit date: | 1997-07-31 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 3D solution structure of copper and silver-substituted yeast metallothioneins.
FEBS Lett., 379, 1996
|
|
1AUX
 
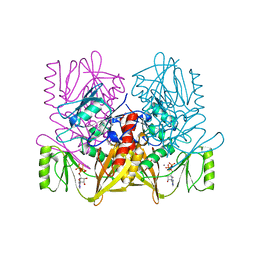 | |
8W0U
 
 | | Human LCAD complexed with Acetoacetyl Coenzyme A | | Descriptor: | ACETOACETYL-COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, Long-chain specific acyl-CoA dehydrogenase, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2024-02-14 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for expanded substrate specificities of human long chain acyl-CoA dehydrogenase and related acyl-CoA dehydrogenases.
Sci Rep, 14, 2024
|
|
8W0Z
 
 | | Human LCAD complexed with Lauric Acid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LAURIC ACID, Long-chain specific acyl-CoA dehydrogenase, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2024-02-14 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for expanded substrate specificities of human long chain acyl-CoA dehydrogenase and related acyl-CoA dehydrogenases.
Sci Rep, 14, 2024
|
|
8W0T
 
 | | Human LCAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Long-chain specific acyl-CoA dehydrogenase, mitochondrial | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2024-02-14 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for expanded substrate specificities of human long chain acyl-CoA dehydrogenase and related acyl-CoA dehydrogenases.
Sci Rep, 14, 2024
|
|
8PPW
 
 | | Structure of human PARK7 in complex with GK16S | | Descriptor: | (3~{S})-1-(iminomethyl)-~{N}-pent-4-ynyl-pyrrolidine-3-carboxamide, Parkinson disease protein 7 | | Authors: | Grethe, C, Gersch, M. | | Deposit date: | 2023-07-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | N-Cyanopiperazines as Specific Covalent Inhibitors of the Deubiquitinating Enzyme UCHL1.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8PQ0
 
 | | Structure of human PARK7 in complex with GK16R | | Descriptor: | (3~{R})-3-(pent-4-ynylcarbamoyl)pyrrolidine-1-carboximidothioic acid, Parkinson disease protein 7 | | Authors: | Grethe, C, Gersch, M. | | Deposit date: | 2023-07-10 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | N-Cyanopiperazines as Specific Covalent Inhibitors of the Deubiquitinating Enzyme UCHL1.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
1BZ7
 
 | | FAB FRAGMENT FROM MURINE ASCITES | | Descriptor: | PROTEIN (ANTIBODY R24 (HEAVY CHAIN)), PROTEIN (ANTIBODY R24 (LIGHT CHAIN)) | | Authors: | Kaminski, M.J, Mackenzie, C.R, Mooibroek, M.J, Dahms, T.E.S, Hirama, T, Houghton, A.N, Chapman, P.B, Evans, S.V. | | Deposit date: | 1998-11-06 | | Release date: | 1999-11-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The role of homophilic binding in anti-tumor antibody R24 recognition of molecular surfaces. Demonstration of an intermolecular beta-sheet interaction between vh domains.
J.Biol.Chem., 274, 1999
|
|
1C1X
 
 | | L-PHENYLALANINE DEHYDROGENASE STRUCTURE IN TERNARY COMPLEX WITH NAD+ AND L-3-PHENYLLACTATE | | Descriptor: | ALPHA-HYDROXY-BETA-PHENYL-PROPIONIC ACID, ISOPROPYL ALCOHOL, L-PHENYLALANINE DEHYDROGENASE, ... | | Authors: | Vanhooke, J.L, Thoden, J.B. | | Deposit date: | 1999-07-22 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rhodococcus L-phenylalanine dehydrogenase: kinetics, mechanism, and structural basis for catalytic specificity.
Biochemistry, 39, 2000
|
|
6A01
 
 | | The crystal structure of Mandelate oxidase Y128F with 3,3-difluoro-2,2-dihydroxy-3-phenylpropionic acid | | Descriptor: | 3,3-difluoro-2,2-dihydroxy-3-phenylpropanoic acid, 4-hydroxymandelate oxidase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | Structural and chemical trapping of flavin-oxide intermediates reveals substrate-directed reaction multiplicity.
Protein Sci., 29, 2020
|
|
