2ADT
 
 | | NMR structure of a 30 kDa GAAA tetraloop-receptor complex. | | Descriptor: | 43-MER | | Authors: | Davis, J.H, Tonelli, M, Scott, L.G, Jaeger, L, Williamson, J.R, Butcher, S.E. | | Deposit date: | 2005-07-20 | | Release date: | 2005-07-26 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | RNA Helical Packing in Solution: NMR Structure of a 30 kDa GAAA Tetraloop-Receptor Complex
J.Mol.Biol., 351, 2005
|
|
1OMC
 
 | | SOLUTION STRUCTURE OF OMEGA-CONOTOXIN GVIA USING 2-D NMR SPECTROSCOPY AND RELAXATION MATRIX ANALYSIS | | Descriptor: | OMEGA-CONOTOXIN GVIA | | Authors: | Davis, J.H, Bradley, E.K, Miljanich, G.P, Nadasdi, L, Ramachandran, J, Basus, V.J. | | Deposit date: | 1993-04-28 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of omega-conotoxin GVIA using 2-D NMR spectroscopy and relaxation matrix analysis.
Biochemistry, 32, 1993
|
|
2I7Z
 
 | |
2I7E
 
 | |
8E8Q
 
 | | Cryo-EM structure of substrate-free DNClpX.ClpP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ghanbarpour, A, Cohen, S, Davis, J.H, Sauer, R.T. | | Deposit date: | 2022-08-25 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structure of substrate-free DNClpX.ClpP
Nat Commun, 2023
|
|
8E91
 
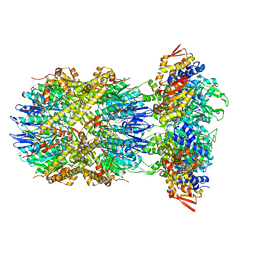 | |
8E7V
 
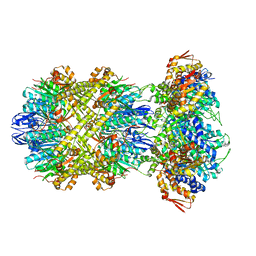 | | Cryo-EM structure of substrate-free DNClpX.ClpP from singly capped particles | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | Authors: | Ghanbarpour, A, Cohen, S, Davis, J.H, Sauer, R.T. | | Deposit date: | 2022-08-24 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of substrate-free DNClpX.ClpP
Nat Commun, 2023
|
|
8ET3
 
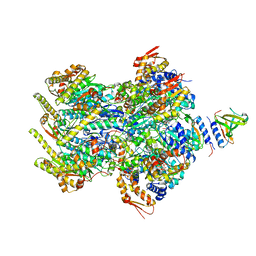 | | Cryo-EM structure of a delivery complex containing the SspB adaptor, an ssrA-tagged substrate, and the AAA+ ClpXP protease | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Ghanbarpour, A, Fei, X, Davis, J.H, Sauer, R.T. | | Deposit date: | 2022-10-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The SspB adaptor drives structural changes in the AAA+ ClpXP protease during ssrA-tagged substrate delivery.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EYT
 
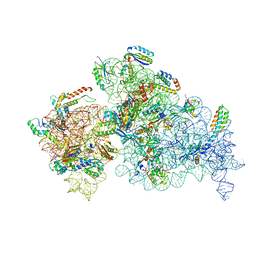 | | 30S_delta_ksgA+KsgA complex | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Sun, J, Kinman, L.F, Jahagirdar, D, Ortega, J, Davis, J.H. | | Deposit date: | 2022-10-28 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | KsgA facilitates ribosomal small subunit maturation by proofreading a key structural lesion.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6UQC
 
 | | Mouse IgG2a Bispecific Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, F, Tsai, J.C, Davis, J.H, West, S.M, Strop, P. | | Deposit date: | 2019-10-18 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Design and characterization of mouse IgG1 and IgG2a bispecific antibodies for use in syngeneic models.
Mabs, 12, 2019
|
|
2AU4
 
 | | Class I GTP aptamer | | Descriptor: | Class I RNA aptamer to GTP, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Carothers, J.M, Davis, J.H, Chou, J.J, Szostak, J.W. | | Deposit date: | 2005-08-26 | | Release date: | 2006-03-28 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an informationally complex high-affinity RNA aptamer to GTP.
Rna, 12, 2006
|
|
