6X5K
 
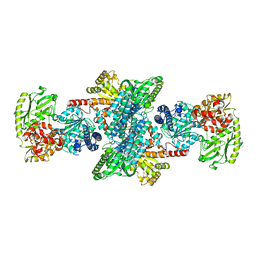 | | Crystal structure of CODH/ACS with carbon monoxide bound to the A-cluster | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, CARBON MONOXIDE, Carbon monoxide dehydrogenase/acetyl-CoA synthase subunit alpha, ... | | Authors: | Cohen, S.E, Wittenborn, E.C, Hendrickson, R, Drennan, C.L. | | Deposit date: | 2020-05-26 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystallographic Characterization of the Carbonylated A-Cluster in Carbon Monoxide Dehydrogenase/Acetyl-CoA Synthase
Acs Catalysis, 10, 2020
|
|
4E49
 
 | |
6V59
 
 | |
3RON
 
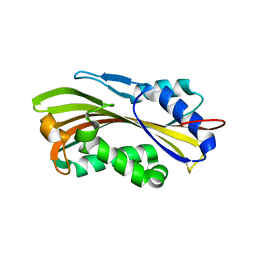 | | Crystal Structure and Hemolytic Activity of the Cyt1Aa Toxin from Bacillus thuringiensis subsp. israelensis | | Descriptor: | Type-1Aa cytolytic delta-endotoxin | | Authors: | Cohen, S, Albeck, S, Ben-Dov, E, Cahan, R, Firer, M, Zaritsky, A, Dym, O, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-04-26 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Cyt1Aa Toxin: Crystal Structure Reveals Implications for Its Membrane-Perforating Function.
J.Mol.Biol., 413, 2011
|
|
4E4A
 
 | |
4E3G
 
 | |
4E3F
 
 | |
4E3D
 
 | |
4E3H
 
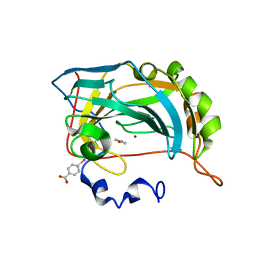 | |
6ONS
 
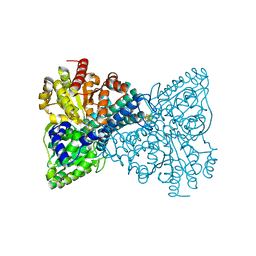 | | Crystal structure of Desulfovibrio vulgaris carbon monoxide dehydrogenase with the D-cluster ligating cysteines mutated to alanines, coexpressed with CooC, as-isolated | | Descriptor: | Carbon monoxide dehydrogenase, FE(4)-NI(1)-S(4) CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Cohen, S.E, Wittenborn, E.C, Drennan, C.L. | | Deposit date: | 2019-04-22 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.485 Å) | | Cite: | Structural insight into metallocofactor maturation in carbon monoxide dehydrogenase.
J.Biol.Chem., 294, 2019
|
|
6NX0
 
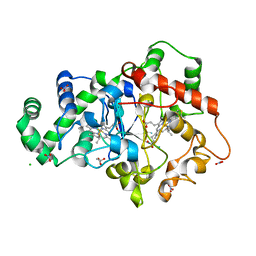 | |
1ODH
 
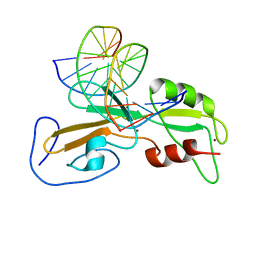 | | Structure of the GCM domain bound to DNA | | Descriptor: | 5'-D(*CP*GP*AP*TP*GP*CP*GP*GP*GP*TP *GP*CP*A)-3', 5'-D(*TP*GP*CP*AP*CP*CP*CP*GP*CP*AP *TP*CP*G)-3', MGCM1, ... | | Authors: | Cohen, S.X, Muller, C.W. | | Deposit date: | 2003-02-19 | | Release date: | 2003-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of the Gcm Domain-DNA Complex: A DNA-Binding Domain with a Novel Fold and Mode of Target Site Recognition
Embo J., 22, 2003
|
|
1LU5
 
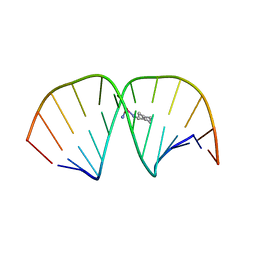 | | 2.4 Angstrom Crystal Structure of the Asymmetric Platinum Complex {Pt(ammine)(cyclohexylamine)}2+ Bound to a Dodecamer DNA Duplex | | Descriptor: | 5'-D(*CP*CP*TP*CP*TP*GP*GP*TP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*AP*CP*CP*AP*GP*AP*GP*G)-3', CIS-(AMMINE)(CYCLOHEXYLAMINE)PLATINUM(II) COMPLEX | | Authors: | Silverman, A.P, Bu, W, Cohen, S.M, Lippard, S.J. | | Deposit date: | 2002-05-21 | | Release date: | 2002-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4-A Crystal Structure of the Asymmetric Platinum Complex {Pt(ammine)(cyclohexylamine)}2+ Bound to
a Dodecamer DNA Duplex
J.Biol.Chem., 277, 2002
|
|
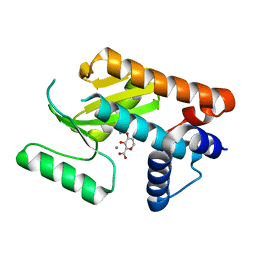
6DCZ
 
 | |
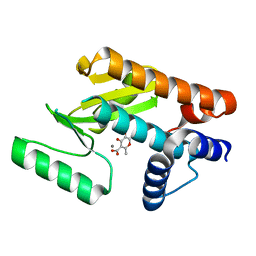
6DCY
 
 | |
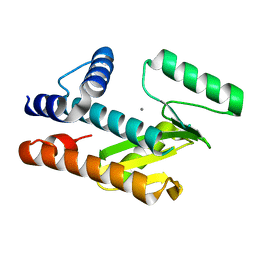
8VGZ
 
 | |
8CTF
 
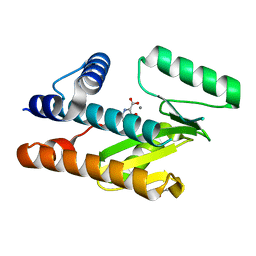 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 3-hydroxy-4-oxo-1,4-dihydropyridine-2-carboxylic acid | | Descriptor: | 3-hydroxy-4-oxo-1,4-dihydropyridine-2-carboxylic acid, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-05-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8DDB
 
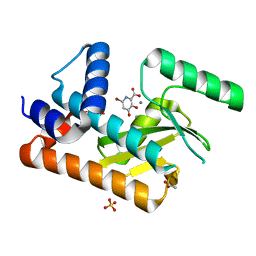 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 4-(benzyloxy)-6-bromo-3-hydroxypicolinic acid | | Descriptor: | 6-bromo-3-hydroxy-4-oxo-1,4-dihydropyridine-2-carboxylic acid, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
9AXC
 
 | | Activated CRAF/MEK heterotetramer from focused refinement of CRAF/MEK/14-3-3 complex | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, GST26/CRAF chimera, N-[3-fluoro-4-({7-[(3-fluoropyridin-2-yl)oxy]-4-methyl-2-oxo-2H-1-benzopyran-3-yl}methyl)pyridin-2-yl]-N'-methylsulfuric diamide | | Authors: | Quade, B, Cohen, S.E, Huang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | The pan-RAF-MEK non degrading molecular glue NST-628 is a potent and brain penetrant inhibitor of the RAS-MAPK pathway with activity across diverse RAS- and RAF-driven cancers.
Cancer Discov, 2024
|
|
9AXA
 
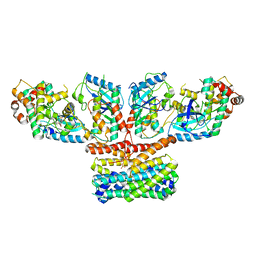 | | CryoEM structure of activated CRAF/MEK/14-3-3 complex with NST-628 | | Descriptor: | 14-3-3 protein sigma, Dual specificity mitogen-activated protein kinase kinase 1, GST26/CRAF chimera, ... | | Authors: | Quade, B, Cohen, S.E, Huang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | The pan-RAF-MEK non degrading molecular glue NST-628 is a potent and brain penetrant inhibitor of the RAS-MAPK pathway with activity across diverse RAS- and RAF-driven cancers.
Cancer Discov, 2024
|
|
8DAL
 
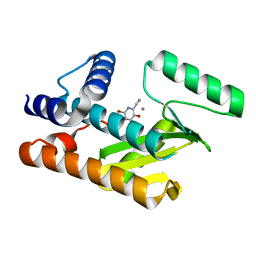 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 4-(benzyloxy)-6-bromo-3-hydroxypicolinonitrile | | Descriptor: | 6-bromo-3-hydroxy-4-oxo-1,4-dihydropyridine-2-carbonitrile, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-06-13 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8DJY
 
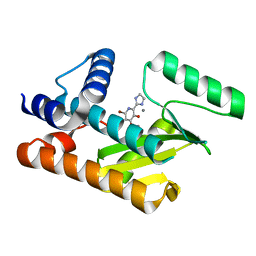 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 6-Bromo-2-(4,5-dihydro-1H-imidazol-2-yl)-3-hydroxypyridin-4(1H)-one | | Descriptor: | (2M)-6-bromo-2-(4,5-dihydro-1H-imidazol-2-yl)-3-hydroxypyridin-4(1H)-one, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-07-01 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8DDE
 
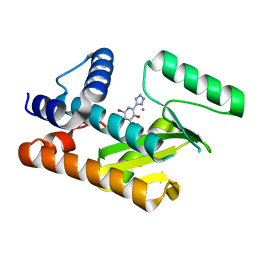 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 4-(benzyloxy)-6-bromo-2-(1H-tetrazol-5-yl) yridine-3-ol | | Descriptor: | (2M)-6-bromo-3-hydroxy-2-(1H-tetrazol-5-yl)pyridin-4(1H)-one, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-06-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8DHN
 
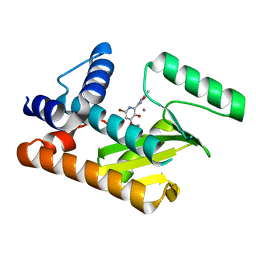 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 6-Bromo-3-hydroxy-2-(5-methyl-1,2,4-oxadiazol-3-yl)pyridin-4(1H)-one | | Descriptor: | (2P)-6-bromo-3-hydroxy-2-(5-methyl-1,2,4-oxadiazol-3-yl)pyridin-4(1H)-one, MANGANESE (II) ION, PA endonuclease | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-06-27 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8DJV
 
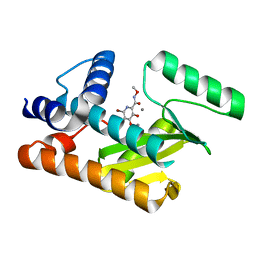 | | The N-terminal domain of PA endonuclease from the influenza H1N1 viral polymerase in complex with 6-Bromo-3-hydroxy-N-methoxy-4-oxo-1,4-dihydropyridine-2-carboxamide | | Descriptor: | 6-bromo-3-hydroxy-N-methoxy-4-oxo-1,4-dihydropyridine-2-carboxamide, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Kohlbrand, A.J, Stokes, R.W, Karges, J, Seo, H, Sankaran, B, Cohen, S.M. | | Deposit date: | 2022-07-01 | | Release date: | 2022-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Carboxylic Acid Isostere Derivatives of Hydroxypyridinones as Core Scaffolds for Influenza Endonuclease Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
