8DD8
 
 | | PI 3-kinase alpha with nanobody 3-142, crosslinked with DSG | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8PYQ
 
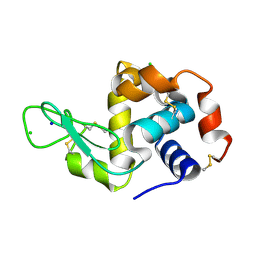 | | 10 micrometer HEWL crystals solved at room-temperature using fixed-target serial crystallography. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Mason, T.J, Carrillo, M, Beale, J.H, Padeste, C. | | Deposit date: | 2023-07-25 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Micro-structured polymer fixed targets for serial crystallography at synchrotrons and XFELs.
Iucrj, 10, 2023
|
|
1NYP
 
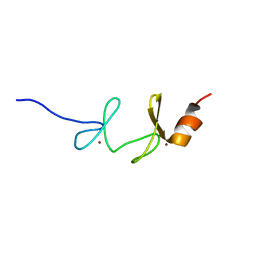 | | 4th LIM domain of PINCH protein | | Descriptor: | PINCH protein, ZINC ION | | Authors: | Velyvis, A, Vaynberg, J, Vinogradova, O, Zhang, Y, Wu, C, Qin, J. | | Deposit date: | 2003-02-13 | | Release date: | 2003-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and functional insights into PINCH LIM4 domain-mediated integrin signaling
Nat.Struct.Biol., 10, 2003
|
|
8PYO
 
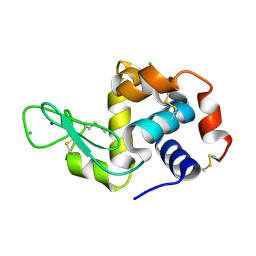 | | 5 micrometer HEWL crystals solved at room-temperature using fixed-target serial crystallography. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Mason, T.J, Carrillo, M, Beale, J.H, Padeste, C. | | Deposit date: | 2023-07-25 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Micro-structured polymer fixed targets for serial crystallography at synchrotrons and XFELs.
Iucrj, 10, 2023
|
|
8Q7C
 
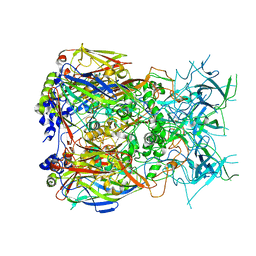 | | Cryo-EM structure of Adenovirus C5 hexon | | Descriptor: | Hexon protein | | Authors: | Zoll, S, Dhillon, A. | | Deposit date: | 2023-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the interaction between adenovirus C5 hexon and human lactoferrin.
J.Virol., 98, 2024
|
|
8PYP
 
 | | 25 micrometer HEWL crystals solved at room-temperature using fixed-target serial crystallography. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Mason, T.J, Carrillo, M, Beale, J.H, Padeste, C. | | Deposit date: | 2023-07-25 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Micro-structured polymer fixed targets for serial crystallography at synchrotrons and XFELs.
Iucrj, 10, 2023
|
|
8DCX
 
 | | PI 3-kinase alpha with nanobody 3-159 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1NOG
 
 | | Crystal Structure of Conserved Protein 0546 from Thermoplasma Acidophilum | | Descriptor: | conserved hypothetical protein TA0546 | | Authors: | Saridakis, V, Sanishvili, R, Iakounine, A, Xu, X, Pennycooke, M, Gu, J, Joachimiak, A, Arrowsmith, C.H, Edwards, A.M, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-16 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The structural basis for methylmalonic aciduria. The crystal structure of archaeal ATP:cobalamin adenosyltransferase.
J.Biol.Chem., 279, 2004
|
|
2RN2
 
 | | STRUCTURAL DETAILS OF RIBONUCLEASE H FROM ESCHERICHIA COLI AS REFINED TO AN ATOMIC RESOLUTION | | Descriptor: | RIBONUCLEASE H | | Authors: | Katayanagi, K, Miyagawa, M, Matsushima, M, Ishikawa, M, Kanaya, S, Nakamura, H, Ikehara, M, Matsuzaki, T, Morikawa, K. | | Deposit date: | 1992-04-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural details of ribonuclease H from Escherichia coli as refined to an atomic resolution.
J.Mol.Biol., 223, 1992
|
|
1NUV
 
 | | The Leadzyme Ribozyme Bound to Mg(H2O)6(II) and Sr(II) at 1.8 A resolution | | Descriptor: | 5'-R(*CP*GP*GP*AP*CP*CP*GP*AP*GP*CP*CP*AP*G)-3', 5'-R(*GP*CP*UP*GP*GP*GP*AP*GP*UP*CP*C)-3', MAGNESIUM ION, ... | | Authors: | Wedekind, J.E, Mckay, D.B. | | Deposit date: | 2003-02-01 | | Release date: | 2003-08-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of the leadzyme at 1.8 A resolution: metal ion binding and the implications
for catalytic mechanism and allo site ion regulation.
BIOCHEMISTRY, 42, 2003
|
|
8Q3O
 
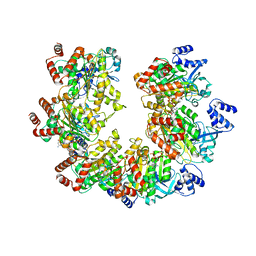 | |
8Q3Q
 
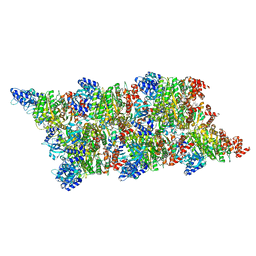 | |
8Q3N
 
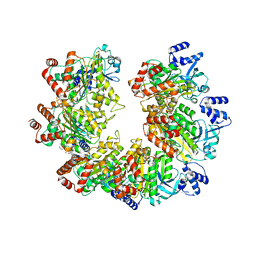 | |
8Q3P
 
 | |
2RMJ
 
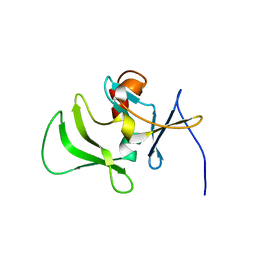 | | Solution structure of RIG-I C-terminal domain | | Descriptor: | Probable ATP-dependent RNA helicase DDX58 | | Authors: | Takahasi, K, Yoneyama, M, Nihishori, T, Hirai, R, Narita, R, Gale Jr, M, Fujita, T, Inagaki, F. | | Deposit date: | 2007-10-23 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nonself RNA-Sensing Mechanism of RIG-I Helicase and Activation of Antiviral Immune Responses
Mol.Cell, 29, 2008
|
|
2SSP
 
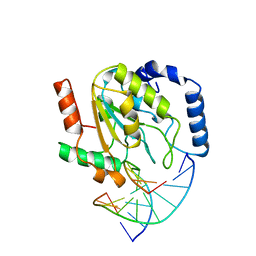 | | LEUCINE-272-ALANINE URACIL-DNA GLYCOSYLASE BOUND TO ABASIC SITE-CONTAINING DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*TP*(AAB)P*AP*TP*CP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE) | | Authors: | Parikh, S.S, Mol, C.D, Slupphaug, G, Bharati, S, Krokan, H.E, Tainer, J.A. | | Deposit date: | 1999-04-28 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
EMBO J., 17, 1998
|
|
8Q92
 
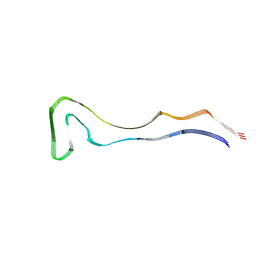 | | P301S Tau Filaments from the Brains of PS19 Transgenic Mouse Line | | Descriptor: | Microtubule-associated protein tau | | Authors: | Schweighauser, M, Murzin, A.G, Macdonald, J, Lavenir, I, Crowther, R.A, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-19 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cryo-EM structures of tau filaments from the brains of mice transgenic for human mutant P301S Tau.
Acta Neuropathol Commun, 11, 2023
|
|
8Q96
 
 | | P301S Tau Filaments from the Brains of Tg2541 Transgenic Mouse Line | | Descriptor: | Isoform Tau-E of Microtubule-associated protein tau, Unknown protein | | Authors: | Schweighauser, M, Murzin, A.G, Macdonald, J, Lavenir, I, Crowther, R.A, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-19 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryo-EM structures of tau filaments from the brains of mice transgenic for human mutant P301S Tau.
Acta Neuropathol Commun, 11, 2023
|
|
2SPL
 
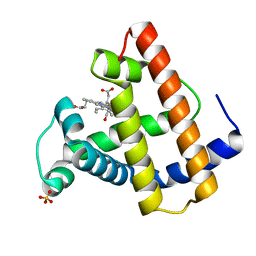 | | A NOVEL SITE-DIRECTED MUTANT OF MYOGLOBIN WITH AN UNUSUALLY HIGH O2 AFFINITY AND LOW AUTOOXIDATION RATE | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Quillin, M.L, Arduini, R.M, Phillips Jr, G.N. | | Deposit date: | 1993-08-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel site-directed mutant of myoglobin with an unusually high O2 affinity and low autooxidation rate.
J.Biol.Chem., 267, 1992
|
|
2UW0
 
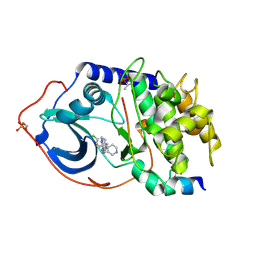 | | Structure of PKA-PKB chimera complexed with 6-(4-(4-(4-Chloro-phenyl) -piperidin-4-yl)-phenyl)-9H-purine | | Descriptor: | 6-{4-[4-(4-CHLOROPHENYL)PIPERIDIN-4-YL]PHENYL}-9H-PURINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Davies, T.G, Donald, A, McHardy, T, Rowlands, M.G, Hunter, L.J, Boyle, R.G, Aherne, G.W, Garrett, M.D, Collins, I. | | Deposit date: | 2007-03-15 | | Release date: | 2007-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rapid Evolution of 6-Phenylpurine Inhibitors of Protein Kinase B Through Structure-Based Design
J.Med.Chem., 50, 2007
|
|
2UW4
 
 | | Structure of PKA-PKB chimera complexed with 2-(4-(5-methyl-1H-pyrazol- 4-yl)-phenyl)-ethylamine | | Descriptor: | 2-[4-(3-METHYL-1H-PYRAZOL-4-YL)PHENYL]ETHANAMINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Davies, T.G, Saxty, G, Woodhead, S.J, Berdini, V, Verdonk, M.L, Wyatt, P.G, Boyle, R.G, Barford, D, Downham, R, Garrett, M.D, Carr, R.A. | | Deposit date: | 2007-03-19 | | Release date: | 2007-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of Inhibitors of Protein Kinase B Using Fragment-Based Lead Discovery
J.Med.Chem., 50, 2007
|
|
2UW5
 
 | | Structure of PKA-PKB chimera complexed with (R)-2-(4-chloro-phenyl)- 2-(4-1H-pyrazol-4-yl)-phenyl)-ethylamine | | Descriptor: | (2R)-2-(4-CHLOROPHENYL)-2-[4-(1H-PYRAZOL-4-YL)PHENYL]ETHANAMINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Davies, T.G, Saxty, G, Woodhead, S.J, Berdini, V, Verdonk, M.L, Wyatt, P.G, Boyle, R.G, Barford, D, Downham, R, Garrett, M.D, Carr, R.A. | | Deposit date: | 2007-03-19 | | Release date: | 2007-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Identification of Inhibitors of Protein Kinase B Using Fragment-Based Lead Discovery
J.Med.Chem., 50, 2007
|
|
2UVX
 
 | | Structure of PKA-PKB chimera complexed with 7-azaindole | | Descriptor: | 1H-PYRROLO[2,3-B]PYRIDINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Davies, T.G, Donald, A, McHardy, T, Rowlands, M.G, Hunter, L.J, Boyle, R.G, Aherne, G.W, Garrett, M.D, Collins, I. | | Deposit date: | 2007-03-15 | | Release date: | 2007-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rapid Evolution of 6-Phenylpurine Inhibitors of Protein Kinase B Through Structure-Based Design
J.Med.Chem., 50, 2007
|
|
2UW7
 
 | | Structure of PKA-PKB chimera complexed with 4-(4-chloro-phenyl)-4-(4- (1H-pyrazol-4-yl)-phenyl)-piperidine | | Descriptor: | 4-(4-CHLOROPHENYL)-4-[4-(1H-PYRAZOL-4-YL)PHENYL]PIPERIDINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Davies, T.G, Saxty, G, Woodhead, S.J, Berdini, V, Verdonk, M.L, Wyatt, P.G, Boyle, R.G, Barford, D, Downham, R, Garrett, M.D, Carr, R.A. | | Deposit date: | 2007-03-19 | | Release date: | 2007-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Inhibitors of Protein Kinase B Using Fragment-Based Lead Discovery
J.Med.Chem., 50, 2007
|
|
2VJB
 
 | | Torpedo Californica Acetylcholinesterase In Complex With A Non Hydrolysable Substrate Analogue, 4-Oxo-N,N,N- Trimethylpentanaminium - Orthorhombic space group - Dataset D at 100K | | Descriptor: | (4R)-4-HYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Colletier, J.P, Bourgeois, D, Fournier, D, Silman, I, Sussman, J.L, Weik, M. | | Deposit date: | 2007-12-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Shoot-and-Trap: Use of Specific X-Ray Damage to Study Structural Protein Dynamics by Temperature-Controlled Cryo-Crystallography.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
